Abstract
Hundreds of species of plant viruses, many of them economically important, are transmitted by noncirculative vector transmission (acquisition by attachment of virions to vector mouthparts and inoculation by subsequent release), but virus receptors within the vector remain elusive. Here we report evidence for the existence, precise location, and chemical nature of the first receptor for a noncirculative virus, cauliflower mosaic virus, in its insect vector. Electron microscopy revealed virus-like particles in a previously undescribed anatomical zone at the extreme tip of the aphid maxillary stylets. A novel in vitro interaction assay characterized binding of cauliflower mosaic virus protein P2 (which mediates virus–vector interaction) to dissected aphid stylets. A P2-GFP fusion exclusively labeled a tiny cuticular domain located in the bottom-bed of the common food/salivary duct. No binding to stylets of a non-vector species was observed, and a point mutation abolishing P2 transmission activity correlated with impaired stylet binding. The novel receptor appears to be a nonglycosylated protein deeply embedded in the chitin matrix. Insight into such insect receptor molecules will begin to open the major black box of this scientific field and might lead to new strategies to combat viral spread.
Keywords: aphid, receptor
Nearly all plant viruses that cause extensive agricultural damage use specific vectors to spread between hosts. The most common vectors are arthropods, especially aphids (1), and the most widely adopted strategy for virus–vector interaction is noncirculative transmission, in which the virus is taken up by a vector feeding on an infected plant, adsorbed somewhere on the cuticle lining the inner part of the feeding apparatus, and subsequently released to inoculate a new host plant. The viral components involved in this interaction are relatively well established, in particular for the genus Cucumovirus, where domains of the coat protein directly recognize unknown retention sites in the vector mouthparts (capsid strategy), and for the genera Potyvirus and Caulimovirus, where a nonstructural viral protein, HC (helper component), creates the link between virion and vector (helper strategy) (reviewed in refs. 2 and 3). However, no putative binding sites for viral components in the insect vector have ever been chemically characterized or even precisely localized. This question is of major importance, because numerous noncirculative viruses may use the same vector attachment sites, and identification of putative receptor molecules could lead to new strategies to combat viral spread.
A distinguishing feature of noncirculative transmission is that several virus species can be transmitted by the same vector, and, conversely, several vector species can transmit the same virus. Hence, although some degree of noncirculative virus/vector specificity exists (4, 5), it is often so broad that the very existence of actual viral receptors remains questionable, because their existence has never been directly demonstrated.
Here we report evidence for the existence, precise location, and chemical nature of the receptor for a noncirculative virus, cauliflower mosaic virus (CaMV), in its insect vector. A novel in vitro system allowed rapid visualization of the interaction between dissected aphid stylets and the HC of CaMV. The CaMV retention sites are concentrated exclusively in a tiny and previously unknown anatomical zone located at the extreme tip of the aphid maxillary stylets. Virus/vector binding at this specific zone is mandatory for successful CaMV transmission. Pretreatment of dissected stylets with various chemicals and enzymes demonstrated that the molecule used by CaMV as a specific receptor for vector transmission is a nonglycosylated protein deeply embedded in the chitin matrix.
Results
Localization of CaMV Retention Sites in Insect Stylets.
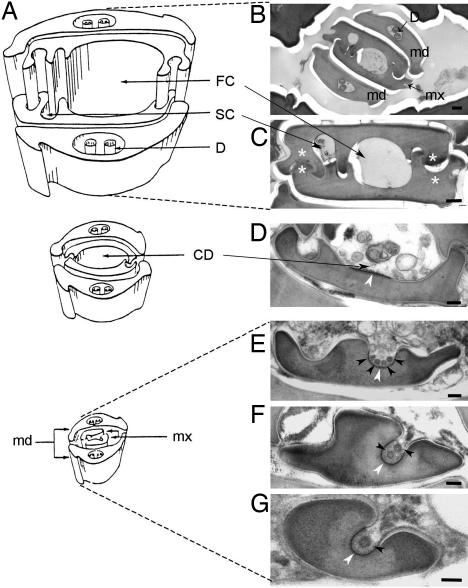
We first investigated, using transmission electron microscopy (TEM), CaMV retention sites in the stylets of the aphid Brevicoryne brassicae fed on infected plants. B. brassicae is a highly efficient CaMV vector (5) and probably retains virus particles longer than other aphid species (6). Examination of the anterior alimentary track beyond the stylets did not detect CaMV-like particles anywhere in the foregut (data not shown). In contrast, in four of six aphids analyzed, spherical virus-like particles (VLPs) were observed in the common food/salivary duct (see Fig. 1A for anatomy of aphid mouthparts) at the extreme tip of the maxillary stylets (Fig. 1 E–G). Because several identical VLPs with an empty central cavity and a mean diameter of 50 nm were often found side by side, they most likely correspond to CaMV particles for which the exact same features have previously been reported (7) [for further evidence of CaMV particles at this location, see supporting information (SI) Fig. 4]. VLPs were never seen in the upper differentiated food or salivary canals of the maxillary stylets (Fig. 1 B and C) or anywhere in the mandibular stylets (data not shown). Notably, all VLPs were localized close to a particular electron-dense area of the surface cuticle. This electron-dense region totally surrounded the lumen of the narrow duct at the distal extremity of individual maxillary stylets (Fig. 1 F and G), lined the bottom-bed of the wider part of the common food/salivary duct where the interlocking ridges and grooves start to differentiate (Fig. 1 D and E), and ended before the bifurcation of the common duct into differentiated canals (Fig. 1 B and C). This anatomical feature of the cuticle surface, which was also consistently observed in aphids fed on noninfected plants (SI Fig. 5), has never been described before by entomologists and is therefore totally enigmatic at this point.
Fig. 1.
CaMV-like particles are observed in the common food/salivary duct located at the tip of the maxillary stylets of aphid vectors. (A) Schematic representation of the stylets bundle anatomy of M. persicae. [Reproduced with permission from ref. 38 (Copyright 1974, Wiley–Blackwell, Oxford, U.K.).] (B–G) Electron micrographs of cross sections of the stylets bundle of a B. brassicae aphid fed on a CaMV-infected plant. Mandibular stylets (md) are located on the outside of the bundle, protecting the maxillary stylets (mx), and interlocked via a complex inner architecture (B). Besides interlocking structures (white asterisks), the inner architecture of maxillary stylets is delimiting a large food canal (FC) and a narrow salivary canal (SC) along the several hundred micrometers of the stylet's length (C). In the ≈5-μm distal extremity of the maxillary stylets, a different inner architecture drives a fusion of FC and SC into single common duct (CD), and the interlocking ridges and grooves progressively disappear as the CD deepens and reduces in diameter (D–G). VLPs are indicated by black arrowheads. The electron-dense area of the cuticle surface distinguishable in the bottom-bed of the CD is indicated by white arrowheads. D, dendrite. (Scale bars: 200 nm in B and 100 nm C–G.)
The impracticality of TEM for routine analyses, and the impossibility of chemically treating stylets of live aphids before CaMV acquisition without affecting their probing behavior, prompted the development of an in vitro virus–vector interaction assay using dissected stylets. The CaMV protein P2 (the HC of CaMV) has long been demonstrated to be the viral product primarily involved in vector recognition (8); thus, a P2-GFP fusion was used to probe the putative receptor at the tip of the maxillary stylets.
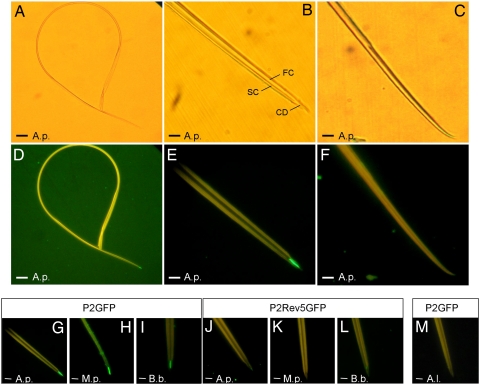
In vitro interaction experiments were carried out with three efficient CaMV vector species: B. brassicae, Myzus persicae, and the pea aphid Acyrthosiphon pisum, with particular focus on the latter because of the ongoing sequencing project of this insect genome (9). A total of 476 A. pisum stylets (324 maxillary and 152 mandibular) was examined for P2-GFP binding. Every maxillary stylet observed showed a sharp fluorescent labeling at its extremity (Fig. 2D, E, and G), whereas no mandibular stylet was ever labeled (Fig. 2F). Consistent with the TEM results, labeling was very specifically restricted to a small distal portion (≈5 μm) of the maxillary stylets, representing <1% of their total length of >700 μm (Fig. 2 A and D) and located downstream of the point where food and salivary canals fuse into a single common duct (Fig. 2 B and E). The area revealed by P2-GFP fluorescence coincides with the location of VLPs observed by TEM and overlaps the electron-dense region of the cuticle surface (Fig. 1).
Fig. 2.
In vitro interaction assay between dissected aphid stylets and P2-GFP. (A–F) Stylets incubated with P2-GFP were observed with bright-field (A–C) or epifluorescence (D–F) microscopy to detect bound P2-GFP. The food (FC) and salivary (SC) canals and the common duct (CD) are indicated in B. Images of the entire stylets (A and D) show that P2-GFP binds exclusively to the maxillary stylet tips. Higher magnification reveals that P2 label is restricted to the food/salivary common duct of the maxillary stylets (B and E). The mandibular stylets (C and F) are never labeled. (G–M) Specificity of the interaction between P2-GFP and maxillary stylets. P2-GFP binds to maxillary stylets from vector species A. pisum (A.p.), M. persicae (M.p.), and B. brassicae (B.b.) (G–I), whereas P2Rev5-GFP, a fusion with a nontransmissible P2 mutant, does not (J–L). (M) The non-vector A. lactucae does not bind P2-GFP. (Scale bars: 25 μm in A and D and 5 μm in B, C, and E–M.)
Specificity of binding was confirmed in several ways: (i) the same results were obtained with M. persicae and B. brassicae (Fig. 2 H and I), (ii) GFP alone did not label stylet tips (SI Fig. 4), (iii) the possibility of artifactual binding of the P2-GFP fusion to stylets was eliminated by detecting the same interaction by immunofluorescence using native P2 (SI Fig. 4), (iv) virions incubated with native P2 were shown to precisely colocalize at the stylet tip (SI Fig. 4), and, (v) as described below, binding of P2 to this restricted stylet area precisely correlates with successful virus transmission.
P2 Binding at the Tip of the Maxillary Stylets Is Required for Aphid Transmission of CaMV.
To confirm the biological significance of the observed P2–maxillary stylet binding, the in vitro interaction between efficient vector species and a defective P2, and between non-vector species and a functional P2, were evaluated.
The mutant protein P2Rev5 (Q to Y substitution at amino acid 6 of P2) is defective in CaMV transmission with all aphid species tested, including B. brassicae and M. persicae, while retaining all other biological properties of wild-type P2 (5). It was proposed that P2Rev5 might be impaired in binding to aphid stylets, a hypothesis not previously testable. After incubation of a P2Rev5-GFP fusion with dissected stylets of A. pisum, M. persicae, and B. brassicae, no labeling was observed on 15 maxillary stylets from M. persicae (Fig. 2K), and only barely detectable P2rev5-GFP signals were seen in six of 10 A. pisum stylets (Fig. 2J) and in 14 of 38 B. brassicae stylets (Fig. 2L). We therefore conclude that the lack of aphid transmission activity of P2Rev5 is due to a defect in stylet binding. The extremely weak interactions inconstantly observed with B. brassicae and A. pisum are obviously below the threshold required for efficient transmission.
Reciprocally, our in vitro binding assay with functional P2-GFP can be used as a tool to predict whether or not an aphid species can transmit CaMV. Although poorly efficient CaMV vector species have been reported, none of >30 aphid species tested thus far are strict CaMV nontransmitters (5, 10–12). While surveying P2-GFP binding to additional aphid species, a total absence of labeling was observed on 31 maxillary stylets from Acyrthosiphon lactucae (Fig. 2M). Consistently, no transmission of CaMV was observed with this species (0/96 test plants), whereas efficient transmission by A. pisum, M. persicae, and B. brassicae was recorded in parallel experiments with rates of 32.3%, 50%, and 66.3%, respectively. Monitoring of A. lactucae using electrical penetration graph control revealed no evidence of any behavioral difference compared with M. persicae that could explain its failure in CaMV transmission (SI Table 1). This report therefore describes A. lactucae as the first species that is a non-vector of CaMV because of the absence of retention sites compatible with its HC (the protein P2).
Biochemical Characterization of the CaMV Receptor.
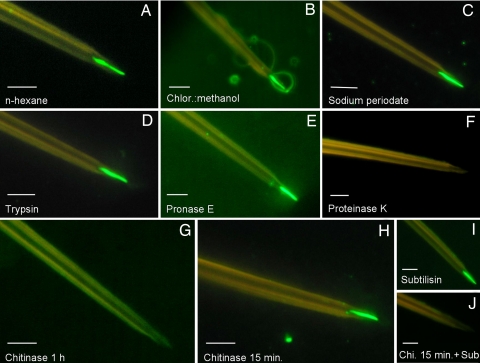
Dissected stylets of A. pisum were submitted to different chemical or enzymatic treatments before P2-GFP in vitro binding assays. Solvent treatments with n-hexane or chloroform:methanol, to extract polar or nonpolar lipids, respectively, did not interfere with subsequent P2-GFP binding (Fig. 3 A and B), ruling out a possible lipidic nature of the CaMV receptor. Exposure of stylets to a range of proteases demonstrated that whereas trypsin, pronase E, and subtilisin had no detectable effect (Fig. 3 D, E, and I), pretreatment with proteinase K totally abolished subsequent P2-GFP attachment on the retention sites (Fig. 3F), indicating that the receptor molecules are, at least partially, proteinaceous. Sodium metaperiodate treatments of various stringencies failed to abolish or reduce the strong P2-GFP labeling of the tip of maxillary stylets (Fig. 3C), suggesting that the protein receptor of CaMV is not glycosylated.
Fig. 3.
Effect of chemical and biochemical pretreatments on binding of P2-GFP to maxillary stylets. Stylets were pretreated with the compounds indicated before P2-GFP incubation and observation by epifluorescence microscopy. (A and B) Extraction of polar lipids (A) and nonpolar lipids (B) with n-hexane and chloroform:methanol, respectively. (C) Sodium periodate oxidization of carbohydrate compounds: all treatments described in Materials and Methods gave identical images. (D–F) Digestion with various proteinases as indicated. (G–J) Effect of pretreatment with chitinase with or without subtilisin. (G) Long chitinase digestion alone. (H) Short chitinase digestion alone. (I) Subtilisin digestion alone (control). (J) Short chitinase digestion followed by subtilisin digestion. (Scale bars: 5 μm.)
Because the proteases used in this study did not have equivalent destructive effects on the receptor, the logical hypothesis that most proteolytic sites are embedded in the chitin matrix and hence are protected from enzymatic cleavage was tested by thoroughly or partially digesting the stylets with chitinase, alone or in combination with protease treatments. A 1-h treatment with chitinase alone totally abolished P2-GFP binding (Fig. 3G and SI Table 2), suggesting that the receptor is lost if the surface of the chitin matrix is extensively degraded. In contrast, limited 15-min chitinase-only exposures retained the receptor accessible to P2-GFP (Fig. 3H and SI Table 2) but rendered it susceptible to subtilisin cleavage (Fig. 3J and SI Table 2), whereas subtilisin was ineffective on non-chitinase-treated stylets (Fig. 3I and SI Table 2).
Discussion
Despite its potential importance in the design of new disease control strategies, the identification of receptors of noncirculative viruses in their insect vectors represents one of the most significant pieces of missing information in the field of virus–vector molecular interactions, for both virologists and entomologists. The complex and largely unresolved composition and structure of the insect cuticle, together with the technical difficulties involved in extracting cuticle components and in the biochemical analyses of live insects, go some way toward explaining the very limited progress in receptor characterization, or even in determining their location in the vectors' anterior alimentary tract. The characterization of the CaMV receptor reported here represents an unprecedented advance toward opening this “black box” by definitively establishing the existence, the precise location, and the biochemical nature of the receptor of a noncirculative virus in its insect vector.
Previous attempts at localizing attachment sites of noncirculative virus species in various vectors only poorly defined the anatomical regions involved in the anterior alimentary tract. The best documented example is certainly that of the genus Potyvirus, where a combination of light and electron microscopy revealed transmissible complexes of tobacco etch virus and tobacco vein mottling virus apparently randomly distributed along the maxillary food canal, sometimes throughout its entire length, sometimes limited to its distal and/or central portions (13–15). A specific correlation between these various distribution patterns and the efficiency of vector transmission could not be investigated, and retention in the short distal common duct, as reported here for CaMV, was not specifically assessed.
Species belonging to the genera Potyvirus and Cucumovirus are retained in an infectious form for only a few minutes in their aphid vectors, whereas viruses in other genera transmitted by aphids or distinct sap-feeding insects can be retained for longer periods extending up to a few hours. The former and latter virus types are referred to as nonpersistent and semipersistent, respectively (16), and are considered as subcategories within the noncirculative transmission mode (17). Virus particles from two semipersistent waikavirus species, transmitted by aphids and leafhoppers, respectively, were observed by TEM not only in the stylets but also higher up in the foregut, encompassing the cibarial pump and the pharynx (18, 19). From this somewhat limited experimental basis, the idea that nonpersistent viruses would be “stylet-borne,” whereas semipersistent viruses would be “foregut-borne,” has developed and become surprisingly prominent in the literature (11, 20–23). The results reported here break this rule by demonstrating that receptors located at the extreme tip of the maxillary stylets are sufficient to mediate transmission of a typical semipersistent virus, i.e., CaMV (12), thus questioning whether any qualitative difference in virus–vector interaction actually exists between non- and semipersistent subcategories.
The observations that various solvents did not extract the receptor protein, that it was poorly accessible to most proteases tested, that limited chitinase treatment exposed it to subtilisin, and that extensive chitinase treatment totally released it are consistent with the conclusion that the receptor is not simply adsorbed at the cuticle surface but is deeply embedded therein, a feature characterizing it as a cuticular protein. Most proteins extracted from the cuticle of diverse arthropods contain distinct sequence signatures, for example the R&R consensus found in the classes Arachnida, Crustacea, and Insecta (24) (25), including hemipteran insects such as aphids (26, 27). This property might facilitate the future identification and cloning of CaMV receptor gene candidates by allowing screening for such sequence signatures among data sets released by the ongoing A. pisum genome project. A recent proteomic approach, betting on the herein demonstrated possibility that a cuticular protein could act as a receptor for aphid transmission of a noncirculative virus, zucchini yellow mosaic virus (a member of the genus Potyvirus), unfortunately failed to extract any detectable proteins from the stylets and to identify the putative receptor among a number of cuticular proteins extracted from whole aphids (M. persicae) (28).
The maxillary stylets, made of cuticle, thought to be composed mainly of chitin, and containing no cells, are poorly characterized organs (29). The localization of the CaMV receptor in a tiny distal area, ≈5 μm long and <1 μm wide, suggests a timely regulation of stylet protein content during ontogenesis. Indeed, our results exclude the possibility that this protein could be homogeneously distributed within the chitin matrix, being accessible at the surface solely in that specific area, because neither extraction of cuticle lipids nor the removal of the outermost chitin layers by chitinase treatment revealed the receptor in other areas of the stylet. We thus propose that the CaMV receptor is transiently expressed at a very early stage of stylet production and that its expression is shut down before the production of the portion of the stylets where the food and salivary canals will be differentiated. Interestingly, the CaMV receptor is present in maxillary stylets of A. pisum nymphs at stages L3 and L4 (data not shown) and thus is presumably important throughout the insect life cycle; however, no sound hypothesis as to the physiological role of this protein can yet be proposed.
Finally, cucumoviruses and potyviruses are inoculated into healthy plants during the initial phase of the aphid probing process, immediately after insertion of the stylet bundle into the cytoplasm of epidermal and/or mesophyll cells during the short probes used by aphids to assess the suitability of the plant as food (30). Although egestion of minute amounts of the food canal content cannot be totally excluded, ejection of watery saliva into the cell during this step has been experimentally proven (31). Logically, then, viruses should bind regions of the stylets that are in contact with (i) contaminated sap streaming in while the aphid feeds on the infected plant and (ii) saliva flushing out during probing of a new healthy host. Thus, the common duct at the tip of the maxillary stylets is ideally placed to harbor virus receptors. The results reported here provide direct experimental proof of the role of the common duct by delimiting a novel anatomical zone of the stylet where the cuticle surface shows a distinct, denser structure, clearly containing the receptors of CaMV and perhaps those of other noncirculative viruses.
Materials and Methods
Aphid Clones and Test Plants.
All aphid species were reared in an environmental growth chamber at a temperature of 23/18°C and a photoperiod of 14/10 h (day/night). Aphid colonies were maintained on broad bean (for A. pisum), turnip cv. “Just Right” (for B. brassicae), eggplant (for M. persicae), and lettuce (for A. lactucae) plants. Turnips were used as test plants for aphid transmission assays.
Purification of CaMV Particles and Production of Recombinant Proteins.
The aphid-transmissible CaMV isolate Cabb B-JI (32) was propagated on turnip plants by mechanical sap inoculation as described (33). Virus particles were purified according to Plisson et al. (7).
GFP and P2-GFP were expressed in the baculovirus/insect cell system using plasmids p119-GFP and p119-P2-GFP. Plasmid p119-GFP was constructed by insertion of the coding sequence of GFP, excised from pUC-GFP (5) with BamHI/BglII digestion, into p119ad4 (34) at the BglII cloning site. For p119-P2-GFP, the P2 coding region from pGm16-P2 was PCR-amplified by using forward and reverse primers including BamHI and BglII restriction sites at their 5′ and 3′ extremities, respectively, the reverse primer omitting the stop codon of P2 and maintaining the frame for GFP fusion. The PCR fragment was cloned into the BamHI site, just upstream of the GFP coding sequence in pUC-GFP, yielding pUC-P2-GFP. The coding sequence of P2-GFP was then excised and inserted into p119ad4 as described above for GFP. Recombinant baculoviruses expressing GFP and P2-GFP were obtained by cotransfecting p119-GFP or p119-P2-GFP plasmids, together with the AcSLP10 baculovirus vector DNA, as described (35). The p119-P2 and p119-P2Rev5-GFP plasmids, as well as the corresponding recombinant baculoviruses, have been previously described (5).
GFP, P2, P2-GFP, and P2Rev5-GFP proteins were produced in Sf9 cells. Forty-eight hours after infection with the relevant recombinant baculovirus, cells from a 75-cm2 culture flask (Falcon) were harvested, pelleted at 500 × g for 5 min, subjected to a freeze–thaw cycle at −20°C, and resuspended in 1 ml of DB5 buffer (50 mM Hepes, pH 7.0/500 mM LiSO4/0.5 mM EGTA/0.2% wt/vol CHAPS). Fifty-microliter aliquots were frozen at −20°C until use.
Aphid Transmission Assays and Electron Microscopy.
Turnip plants, systemically infected with CaMV Cabb B-JI (3–5 weeks after inoculation), were used as virus source plants. The transmission assays were conducted as described previously (5). The test plants were then transferred to an insect-free growth chamber and visually checked 3 weeks later for symptom development.
Alternatively, after a long acquisition period of 15 h enhancing transmission efficiency (6, 36), B. brassicae aphids were carefully removed from infected leaves and immediately anesthetized with CO2. The anterior part of the head of each aphid, including the labium, was severed in 0.1 M sodium cacodylate buffer (pH 7.2; fixation buffer FB) with a sharp razor blade under a dissecting microscope and transferred immediately into 4% glutaraldehyde FB for 4 h at 4°C. Samples were postfixed in 1% osmium tetraoxide in FB for 1 h at 4°C in the dark, dehydrated in graded series of acetone up to 100%, and embedded in epoxy resin (TAAB 812). Sections 80 nm thick were contrasted with uranyl acetate and lead citrate and observed in a JEOL 100CXII microscope operated at 60–80 kV.
Electrical Penetration Graph Control of Aphid Probing Behavior.
Adult apterous A. lactucae (non-vector) and M. persicae (efficient vector) were attached to a fine gold wire and connected to the input of an electrical penetration graph DC amplifier device as described (6). “Wired” aphids were allowed to probe on CaMV-infected turnip plants for 5 min, and all electrical penetration graph signals were recorded and analyzed with PROBE 3.0 software. A Mann–Whitney U statistical test was applied to the recorded data to reliably rule out significant behavioral differences between the two aphid species (SI Table 2).
P2/Stylet in Vitro Binding Assays.
Aphids of various species were freeze-killed at −20°C for 2 h before dissection and then thawed at room temperature. The stylet bundle of each aphid was freed from the stylet groove of the proboscis under a dissecting microscope (Leica MZ6) by using thin tweezers (no. 5 Dumont). Mandibular and maxillary stylets were carefully separated by using insect pins (0.1 mm; Agar Scientific), transferred to siliconized cover slides (Hampton Research), and heated for 2 h at 50°C for increased adhesion to the slides.
Stylets were preincubated with DB5-0 buffer (DB5 without CHAPS) containing 3% BSA (Roche) for 30 min at room temperature and rinsed twice with DB5-0. Protein binding was allowed to proceed for 2 h at room temperature and in the dark by incubating the cover slides with the baculovirus-infected Sf9 cell suspensions described above, diluted 1/16 in DB5 containing 0.05% CHAPS. Stylets were finally rinsed twice for 5 min in DB5-0. Cover slides with attached stylets were mounted on a microscope slide and observed with an Olympus BX60 microscope equipped for epifluorescence.
Stylet-bound native P2 was revealed by incubation with a primary polyclonal antibody raised against P2 (37), followed by incubation with Alexa Fluor 488-conjugated anti-rabbit IgG (Molecular Probes), diluted 1:1,000 and 1:300, respectively, in TBS buffer (50 mM Tris/200 mM NaCl, pH 7.4) supplemented with 5% wt/vol skim milk powder at room temperature. Three rinses of 5 min with TBS were performed after each incubation period.
Chemical and Enzymatic Pretreatments of Stylets.
Various chemical and enzymatic pretreatments were performed on the cover slide-bound stylets to analyze their effect on the ulterior binding of P2-GFP. In independent experiments the stylets were pretreated as follows: (i) pure n-hexane (Sigma) for 1 h at room temperature; (ii) chloroform/methanol [2:1 (vol/vol)] for 30 min followed by chloroform/methanol [1:2 (vol/vol)] for 10 min and pure methanol for 1 min (all steps at room temperature); (iii) 50 mM sodium periodate (Sigma) in 50 mM sodium acetate buffer (pH 5.5) in the dark for 2 h at room temperature, 2 h at 4°C, or 16 h at 4°C; (iv) 1 mg/ml trypsin (Sigma) in PBS-SDS buffer (4.3 mM Na2HPO4/1.4 mM KH2PO4/137 mM NaCl/2.7 mM KCl, pH 7.3/0.05% wt/vol SDS) for 2 h at 37°C; (v) 1 mg/ml pronase E (Sigma) in PBS-SDS buffer for 2 h at 37°C; (vi) 0.5 mg/ml proteinase K (Roche) in PBS-SDS buffer for 2 h at 37°C; (vii) 1 mg/ml subtilisin A (Sigma) in PBS-SDS buffer for 2 h at 37°C; (viii) 1 unit of chitinase from Streptococcus griseus (Sigma) in 50 mM phosphate buffer (pH 6.0) for either 15 min or 1 h at room temperature. Some of these treatments were also performed in combination as indicated in the text, and all were followed by rinses in DB5-0 before P2-GFP binding assays.
Supplementary Material
Acknowledgments
We thank Yvan Rahbe (Institut National de la Recherche Agronomique, Lyon, France) for providing the LL01 clone of A. pisum, Gérard Labonne for breeding various aphid species, and Caulimovirus & Geminivirus: Transmission & Evolution (CaGeTE) team members for helpful discussions. Seeds of turnip cv. “Just Right” were generously provided by Takii Europe. This work was funded by a CT1 grant from the Institut National de la Recherche Agronomique Département Santé des Plantes et Environnement (France), a JE77 grant from the Institut National de la Recherche Agronomique Scientific Direction Plantes et Produits du Végétal (France), and Grant AGL2003-07532-C03-01 from the Ministerio de Educación y Ciencia of Spain.
Abbreviations
- CaMV
cauliflower mosaic virus
- VLP
virus-like particle
- TEM
transmission electron microscopy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 17905.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706608104/DC1.
References
- 1.Hull R. Matthews' Plant Virology. 4th Ed. San Diego: Academic; 2001. [Google Scholar]
- 2.Pirone TP, Blanc S. Annu Rev Phytopathol. 1996;34:227–247. doi: 10.1146/annurev.phyto.34.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Blanc S. In: Viral Transport in Plants. Waigmann E, Heinlein M, editors. Berlin: Springer; 2007. pp. 1–28. [Google Scholar]
- 4.Wang RY, Powell G, Hardie J, Pirone TP. J Gen Virol. 1998;79:1519–1524. doi: 10.1099/0022-1317-79-6-1519. [DOI] [PubMed] [Google Scholar]
- 5.Moreno A, Hebrard E, Uzest M, Blanc S, Fereres A. J Virol. 2005;79:13587–13593. doi: 10.1128/JVI.79.21.13587-13593.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreno A, Palacios I, Blanc S, Fereres A. Ann Entomol Soc Am. 2005;98:763–769. [Google Scholar]
- 7.Plisson C, Uzest M, Drucker M, Froissart R, Dumas C, Conway J, Thomas D, Blanc S, Bron P. J Mol Biol. 2005;346:267–277. doi: 10.1016/j.jmb.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 8.Drucker M, Froissart R, Hebrard E, Uzest M, Ravallec M, Esperandieu P, Mani JC, Pugniere M, Roquet F, Fereres A, et al. Proc Natl Acad Sci USA. 2002;99:2422–2427. doi: 10.1073/pnas.042587799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauthier JP, Legeai F, Zasadzinski A, Rispe C, Tagu D. Bioinformatics. 2007;23:783–784. doi: 10.1093/bioinformatics/btl682. [DOI] [PubMed] [Google Scholar]
- 10.Broadbent L. In: Agricultural Research Council Report Series. Broadbent L, editor. Cambridge, UK: Cambridge Univ Press; 1957. p. 99. [Google Scholar]
- 11.Kennedy JS, Day MF, Eastop VF. A Conspectus of Aphids as Vectors of Plant Viruses. London: Commonwealth Inst of Entomol; 1962. [Google Scholar]
- 12.Markham PG, Pinner MS, Raccah B, Hull R. Ann Appl Biol. 1987;111:571–587. [Google Scholar]
- 13.Berger PH, Pirone TP. Virology. 1986;153:256–261. doi: 10.1016/0042-6822(86)90028-0. [DOI] [PubMed] [Google Scholar]
- 14.Ammar ED, Järlfors U, Pirone TP. Phytopathology. 1994;84:1054–1060. [Google Scholar]
- 15.Wang RY, Ammuar ED, Thornbury DW, Lopez-Moya JJ, Pirone TP. J Gen Virol. 1996;77:861–867. doi: 10.1099/0022-1317-77-5-861. [DOI] [PubMed] [Google Scholar]
- 16.Sylvester ES. J Econ Entomol. 1956;49:789–800. [Google Scholar]
- 17.Harris KF. In: Aphids as Virus Vectors. Harris KF, Maramorosch K, editors. New York: Academic; 1977. pp. 166–208. [Google Scholar]
- 18.Murant AF, Roberts IM, Elnagar S. J Gen Virol. 1976;31:47–57. [Google Scholar]
- 19.Ammar ED, Nault LR. Phytopathology. 1991;81:444–448. [Google Scholar]
- 20.Nault LR, Ammar ED. Annu Rev Entomol. 1989;34:503–529. [Google Scholar]
- 21.Nault LR. Ann Entomol Soc Am. 1997;90:521–541. [Google Scholar]
- 22.Gray SM, Banerjee N. Microbiol Mol Biol Rev. 1999;63:128–148. doi: 10.1128/mmbr.63.1.128-148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng JC, Falk BW. Annu Rev Phytopathol. 2006;44:183–212. doi: 10.1146/annurev.phyto.44.070505.143325. [DOI] [PubMed] [Google Scholar]
- 24.Willis JH. Am Zool. 1999;39:600–609. [Google Scholar]
- 25.Willis JH, Iconomidou VA, Smith RF, Hamodrakas SJ. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill SS, editors. New York: Elsevier; 2005. pp. 79–110. [Google Scholar]
- 26.Iconomidou VA, Willis JH, Hamodrakas SJ. Insect Biochem Mol Biol. 2005;35:553–560. doi: 10.1016/j.ibmb.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 27.Dombrovsky A, Sobolev I, Chejanovsky N, Raccah B. Comp Biochem Physiol B Biochem Mol Biol. 2007;146:256–264. doi: 10.1016/j.cbpb.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Dombrovsky A, Gollop N, Chen S, Chejanovsky N, Raccah B. J Gen Virol. 2007;88:1602–1610. doi: 10.1099/vir.0.82769-0. [DOI] [PubMed] [Google Scholar]
- 29.Parrish WB. Ann Entomol Soc Am. 1967;60:273–276. [Google Scholar]
- 30.Martin B, Collar JL, Tjallingii WF, Fereres A. J Gen Virol. 1997;78:2701–2705. doi: 10.1099/0022-1317-78-10-2701. [DOI] [PubMed] [Google Scholar]
- 31.Powell G. J Gen Virol. 2005;86:469–472. doi: 10.1099/vir.0.80632-0. [DOI] [PubMed] [Google Scholar]
- 32.Delseny M, Hull R. Plasmid. 1983;9:31–41. doi: 10.1016/0147-619x(83)90029-x. [DOI] [PubMed] [Google Scholar]
- 33.Blanc S, Schmidt I, Kuhl G, Esperandieu P, Lebeurier G, Hull R, Cerutti M, Louis C. Virology. 1993;197:283–292. doi: 10.1006/viro.1993.1589. [DOI] [PubMed] [Google Scholar]
- 34.Hébrard E, Drucker M, Leclerc D, Hohn T, Uzest M, Froissart R, Strub J-M, Sanglier S, van Dorsselaer A, Padilla A, et al. J Virol. 2001;75:8538–8546. doi: 10.1128/JVI.75.18.8538-8546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaabihi H, Ogliastro MH, Martin M, Giraud C, Devauchelle G, Cerutti M. J Virol. 1993;67:2664–2671. doi: 10.1128/jvi.67.5.2664-2671.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palacios I, Drucker M, Blanc S, Leite S, Moreno A, Fereres A. J Gen Virol. 2002;83:3163–3171. doi: 10.1099/0022-1317-83-12-3163. [DOI] [PubMed] [Google Scholar]
- 37.Blanc S, Cerutti M, Usmany M, Vlak JM, Hull R. Virology. 1993;192:643–650. doi: 10.1006/viro.1993.1080. [DOI] [PubMed] [Google Scholar]
- 38.Taylor CE, Robertson WM. J Phytopathol. 1974;80:257–266. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.