Abstract
AAA+ proteins play crucial roles in diverse biological processes via their ATPase-driven remodeling of macromolecular complexes. Here we report our identification of an evolutionarily conserved AAA+ protein, ANCCA/pro2000, endowed with a bromodomain that is strongly induced by estrogen in human breast cancer cells and is a direct target of protooncogene ACTR/AIB1/SRC-3. We found that ANCCA associates directly with estrogen-bound estrogen receptor (ER) α and ACTR. It is selectively recruited, upon estrogen stimulation, to a subset of ERα target genes including cyclin D1, c-myc, and E2F1 and is required for their estrogen-induced expression as well as breast cancer cell proliferation. Further studies indicate that ANCCA binds and hydrolyzes ATP and is critical for recruitment of coregulator CBP and histone hyperacetylation at the ER target chromatin. Moreover, mutations at the ATP binding motifs rendered ANCCA defective as a coactivator in mediating estrogen induction of gene expression. Together, our findings reveal an unexpected layer of regulatory mechanism in hormone signaling mediated by ANCCA and suggest that hormone-induced assembly of transcriptional coregulator complexes at chromatin is a process facilitated by AAA+ ATPase proteins.
Keywords: AAA+ protein, cell cycle, nuclear receptors
Estrogen [17β-estradiol (E2)] signaling through estrogen receptors (ERα and ERβ) plays a pivotal role in normal growth and function of the female reproductive system and in the pathogenesis of the majority of human breast cancers (1, 2). ERs, members of the nuclear hormone receptor superfamily, function primarily through transcriptional control of gene expression, which is facilitated by nuclear coregulator proteins (3–9). E2 induction of genes such as cyclin D1 and c-myc in human breast cancer cells likely involves a coordinated recruitment and assembly of a large number of coregulators (10–13). One model proposed for the assembly is protein–protein interaction-mediated docking. For instance, in addition to interacting with nuclear receptors, members of the p160/SRC family can also associate with other coregulators such as p300/CBP and CARM1 (14). Likewise, targeting of the ATP-dependent chromatin-remodeling complexes and the Mediator complex to hormone-responsive genes appears to involve direct interactions between components of the complex and the receptors (15–17). Evidence has also been presented that even though the coactivators may be sequentially recruited to chromatin they may function interdependently (18), suggesting that hormone-induced recruitment of the different coregulators and/or their assembly at the receptor target gene locus is a facilitated process. However, mechanisms underlying the facilitation are not defined.
AAA+ (ATPases associated with various cellular activities) proteins constitute a large family of evolutionarily conserved enzymes that appear to function by causing conformational changes in their substrate proteins or complexes (19–22). Some of the extensively studied, eukaryotic AAA+ proteins such as p97/VCP, NSF, and dynein have diverse functions including membrane fusion, protein degradation, and microtubule sliding. Others, such as the RFC–PCNA complex and CDC6, are involved in the loading or assembly of multiprotein complexes in DNA replication (19). These proteins contain one or more AAA+ domains that are a subfamily of P-loop ATPases with the Walker A and B motifs. However, they differ from the other ATPases, including the SNF2 family of chromatin-remodeling helicases, in several structural and functional features (20). The most salient feature is that AAA+ proteins usually function in an oligomeric form, where a structural motif called arginine fingers from one subunit constitutes part of the nucleotide-binding site of an adjacent subunit.
Here we report the identification of ANCCA, a previously predicted AAA+ protein with a bromodomain, as an ERα coactivator with expression highly induced by E2. We provide evidence that ANCCA mediates E2-stimulated expression of key cell cycle regulators likely via its ATP-driven protein complex-remodeling function and discuss potential implications for ANCCA in breast cancer.
Results
ANCCA, an Evolutionarily Conserved AAA+ Protein, Is a Direct Target of ACTR and Is Strongly Induced by E2.
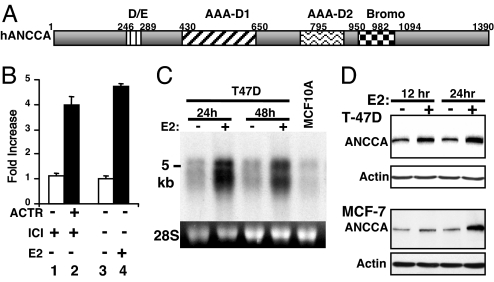
We previously found that overexpression of ACTR promotes breast cancer cell proliferation and anti-E2 resistance. To further understand its functional mechanism, we performed gene expression microarray analysis of breast cancer cells overexpressing ACTR or treated with E2. Both array studies demonstrated up-regulation of an ORF called pro2000 or atad2 with two predicted AAA+ ATPase domains (AAA-D1 and AAA-D2) and a bromodomain. To reflect its function as described below and its link to cancer (below and see Discussion), as well as its recognizable domain feature, we designate this gene as AAA+ nuclear coregulator cancer-associated, or ANCCA (Fig. 1A). Interestingly, ANCCA orthologs with similar AAA+ and bromodomain composition can be found in the current database across the entire eukaryotes [except Drosophila melanogaster; see supporting information (SI) Fig. 7A]. However, their biological functions are largely unknown. Ectopic ACTR induced the expression of ANCCA 4-fold over the control vector in T-47D cells rendered quiescent by anti-estrogen ICI182,780 (Fig. 1B). Likewise, when the cells were treated with E2, ANCCA expression was also strongly induced. E2 induction can be readily detected as early as 6 h after the treatment (SI Fig. 8A). Northern blotting yielded similar results of E2-induced expression of ANCCA mRNA (5-kb and 4.7-kb mRNAs) (Fig. 1C). Notably, the nonmalignant human mammary epithelial cell line MCF-10A expressed a relatively low level of ANCCA. Western blotting detected ANCCA protein with molecular mass of ≈170 kDa in the breast cancer cells, which was highly induced by E2 as early as 12 h after E2 addition (Fig. 1D). Immunofluorescence study revealed ANCCA localized primarily in the nucleus (SI Fig. 8B). ChIP assay with anti-ACTR antibody detected a specific occupancy of ACTR at the predicted transcription start region of ANCCA, indicating that ACTR directly controls ANCCA expression (SI Fig. 8C).
Fig. 1.
ANCCA is a direct target of ACTR and is strongly induced by E2. (A) Schematics of ANCCA subdomains. (B) ANCCA expression in T-47D cells was analyzed by real-time RT-PCR with cells treated with anti-E2 ICI182,780 (10−8 M) for 36 h, then infected with adenovirus vectors for GFP or ACTR (lanes 1 and 2) for 24 h or with cells treated with vehicle or 10−8 M E2 (lanes 3 and 4). (C) Northern blot analysis of ANCCA expression in T-47D cells treated with E2 and MCF-10A cells in regular growth medium. (D) Western blot analysis of ANCCA expression in cells treated with E2 for 12 or 24 h.
ANCCA Is Required for E2-Induced Cell Proliferation and Cell Cycle Progression.
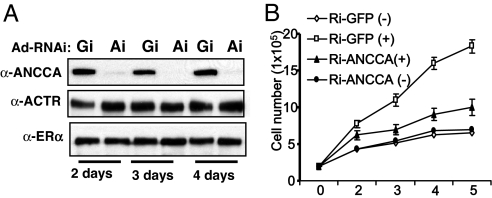
Our finding that ANCCA is regulated by E2 prompted us to examine its function in E2-stimulated cell proliferation. We thus performed RNAi knockdown experiments and assessed the effect on cell proliferation. Adenovirus vector-mediated expression of a shRNA sequence against ANCCA specifically suppressed its expression in T-47D cells with a strong silencing effect observed 2 days after the adeno-vector treatment (Fig. 2A, Gi, GFP-RNAi and Ai, ANCCA-RNAi). Remarkably, knocking down ANCCA was highly inhibitory to E2-stimulated proliferation of T-47D cells (Fig. 2B). Similarly, transfection of MCF-7 cells with a synthetic siRNA targeting a different region of ANCCA mRNA also resulted in strong inhibition of E2-stimulated cell proliferation (SI Fig. 9A), indicating that the effect of ANCCA RNAi on proliferation is not cell-specific or due to a particular siRNA sequence. To assess the potential mechanistic defect underlying the observed effect on cell proliferation, we examined the cell cycle profile. MCF-7 cells transfected with siRNA were treated with E2 for 24 h before harvesting for flow cytometry analysis. As expected from a previous study that a primary effect of E2 is to stimulate G1–S transition, in control siRNA transfected cells, E2 increased S-phase cells from 15% to ≈48% (thus 33% net increase of cell population in S phase). In contrast, in ANCCA siRNA transfected cells, E2 caused an ≈14% increase in S phase (SI Fig. 9B). These results indicate that depletion of ANCCA strongly impedes E2-dependent cell cycle progression from G1 to S. Taken together, these results suggest that ANCCA plays a critical role in E2-stimulated proliferation of human breast cancer cells and that one of the mechanisms is promoting G1–S transition.
Fig. 2.
ANCCA knockdown inhibits E2-stimulated cell proliferation and G1–S progression. (A) T-47D cells being deprived of hormone were infected with equal titers of adeno-shRNA specifically targeting GFP (Gi) or ANCCA (Ai) and harvested at different days after E2 treatment for Western blot analysis. (B) T-47D cells being deprived of hormone were infected with the adeno-shRNAs as in A and 2 days later treated with 10−8 M E2 (+) or vehicle (−) for different days before being harvested for cell numeration.
ANCCA Is Recruited to Selective ERα Target Genes and Is Required for Their Induced Expression.
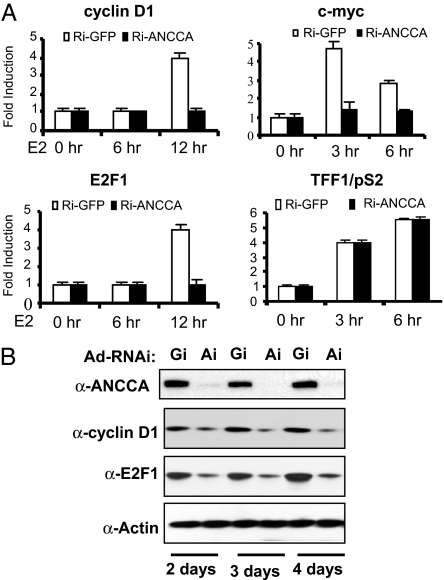
Previous studies demonstrated that E2-stimulated breast cancer cell proliferation involves ERα-mediated direct control of the expression of key cell cycle regulatory genes (23–26). Because ANCCA is a nuclear protein with a bromodomain, which is often found in chromatin/transcriptional regulators, it is possible that ANCCA plays a role in transcriptional regulation mediated by ERα. As shown in Fig. 3A, depletion of ANCCA almost completely blocked E2 induction of cyclin D1, c-myc, and E2F1 mRNA. Remarkably, E2-induced pS2 and cathepsin D transcripts were not significantly affected (Fig. 3A and data not shown). Consistent with the mRNA analysis, Western blotting demonstrated that ANCCA depletion resulted in a significant decrease of cyclin D1 and E2F1 proteins (Fig. 3B).
Fig. 3.
ANCCA depletion inhibits selectively E2-stimulated expression of cyclin D1, c-myc, and E2F1. T-47D cells deprived of hormone were infected with adeno-shRNA targeting GFP (Gi) or ANCCA (Ai), treated with E2 for the indicated hours, and harvested for quantitative mRNA analysis (A) or harvested at different days after E2 treatment for Western blot analysis (B). Fold induction was obtained by comparing real-time RT-PCR data from cells not treated with E2 and cells treated for 6 or 12 h.
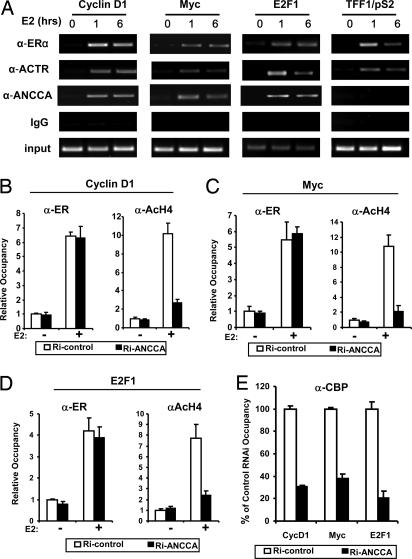
To examine whether ANCCA is directly involved in ER-mediated control of transcription we performed ChIP analysis. As shown previously (10–12), treating cells with E2 induced a robust recruitment of ERα and coactivator ACTR at the ER target gene promoter region of cyclin D1, c-myc, E2F1, and pS2 (Fig. 4A). Strikingly, in the same ChIP experiment, anti-ANCCA antibody detected strong E2-induced recruitment/occupancy of ANCCA at 1 h and 6 h of E2 treatment at the same regions of cyclin D1, E2F1, and c-myc genes that ERα and ACTR occupy. However, on pS2 promoter, no significant recruitment/occupancy of ANCCA was detected under the same condition (Fig. 4A, far right), which is consistent with the data that ANCCA depletion did not significantly affect pS2 gene expression. Together, these results strongly suggest that ANCCA is directly involved in E2 induction of a specific subset of ERα target gene expression.
Fig. 4.
ANCCA is selectively recruited to a subset of ERα target genes, and its knockdown suppresses E2-induced coregulator recruitment and histone hyperacetylation. (A) Hormone-deprived MCF-7 cells were either untreated or treated with E2 for 1 or 6 h before being harvested for ChIP assay with indicated antibodies. Precipitated DNA was analyzed for protein occupancy at indicated promoters by semiquantitative PCR. The entire experiment was repeated three times. (B–D) MCF-7 cells were transfected with either ANCCA siRNA or control siRNA, maintained in hormone-depleted medium for 3 days, and then treated with E2 for 1 h before being harvested for ChIP assays with the indicated antibodies. Relative occupancy was determined by quantifying ChIP PCR products obtained from three experiments, with data from cells transfected with control siRNA and without E2 treatment set at 1. Each bar represents the average from three experiments. (E) E2-induced CBP occupancy at promoters indicated at the bottom was analyzed by ChIP assay as in A–C. No significant difference was observed between non-E2-treated cells that were transfected with either control siRNA or ANCCA siRNA.
ANCCA Is Required for E2 Induction of CBP Recruitment and Chromatin Histone Hyperacetylation.
To further investigate the functional mechanism of ANCCA, we first examined the effect of its depletion on two of the fundamental activities at chromatin in ERα-mediated gene expression, i.e., E2-induced ERα recruitment to target genes and histone hyperacetylation. Notably, depletion of ANCCA had no significant effect on E2-induced ERα recruitment at the ERα target genes examined (Fig. 4 B–D Left). In contrast, ANCCA depletion dramatically suppressed E2-induced histone H4 acetylation at the proximal promoter region of cyclin D1, c-myc, and E2F1 (Fig. 4 B–D Right). Because CBP play crucial roles in E2-induced histone hyperacetylation, we next examined whether CBP occupancy is affected by ANCCA depletion. Strikingly, its depletion strongly inhibited E2-induced CBP occupancy on all three promoters (a 3- to 5-fold decrease) (Fig. 4E). Together, these results suggest that, although ANCCA may not be critical for ERα recruitment to its target genes, it plays an important role in the recruitment or assembly of ERα–CBP complex at the chromatin and hence the histone modifications mediated by the complex.
ANCCA Directly Interacts with ERα and ACTR.
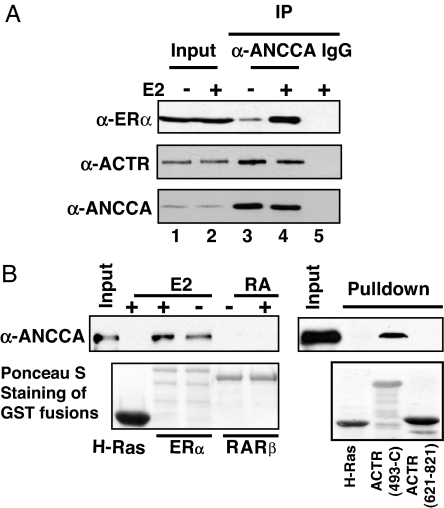
To gain further insight into the mechanism of ANCCA action, we next asked whether ANCCA physically associates with ERα or coactivators such as ACTR. Therefore, we first performed coimmunoprecipitation experiments with nuclear extracts from MCF-7 cells treated with or without E2. Indeed, anti-ANCCA antibody detected strong association between ANCCA and ERα, as a significant portion (>15% of input; compare lanes 2 and 4 of Fig. 5A) of endogenous ERα was coprecipitated by the anti-ANCCA antibody. Importantly, its association with ERα is highly enhanced by E2 (Fig. 5A, compare lane 3 with lane 4). Interestingly, a strong association between ANCCA and ACTR was also detected by the coimmunoprecipitation. To examine whether ANCCA directly interacts with ERα and ACTR, we performed a GST pull-down assay with purified recombinant proteins (Fig. 5B). We found that GST-ERα, but not GST (data not shown) or irrelevant GST-Ras, was able to pull down ANCCA, indicating specific and direct interactions between ERα and ANCCA. Notably, the interaction is significantly enhanced by E2. Interestingly, GST-ACTR protein (amino acid 493-C with N terminus truncated), but not GST-ACTR-RID (receptor interaction domain, amino acids 621–821) specifically pulled down ANCCA, suggesting that ACTR may interact with ANCCA and ERα via distinct domains.
Fig. 5.
ANCCA directly associates with ERα and ACTR. (A) Endogenous ANCCA, ERα, and ACTR protein complexes were analyzed by coimmunoprecipitation assay. Nuclear extracts from MCF-7 cells treated with E2 for 45 min were immunoprecipitated with anti-ANCCA antibody or normal rabbit IgG and blotted by indicated antibodies at the left. Input represents 15% of the extract used in the coimmunoprecipitation. (B) Direct interactions between ANCCA and ERα or ACTR were analyzed by GST pull-down assay using purified proteins. Flag-ANCCA protein (≈100 ng) was incubated with GST-H-Ras (as negative control), GST-ERα, GST-RARβ, or GST-ACTR immobilized on glutathione beads in the presence or absence of E2 (10−7 M) or all-trans retinoic acid (10−7 M). ANCCA retained on the beads was detected by Western blotting. GST fusions were stained with Ponceau S. Input represents 20% of ANCCA proteins used in the individual pull-down.
ANCCA Possesses ATPase Activity.
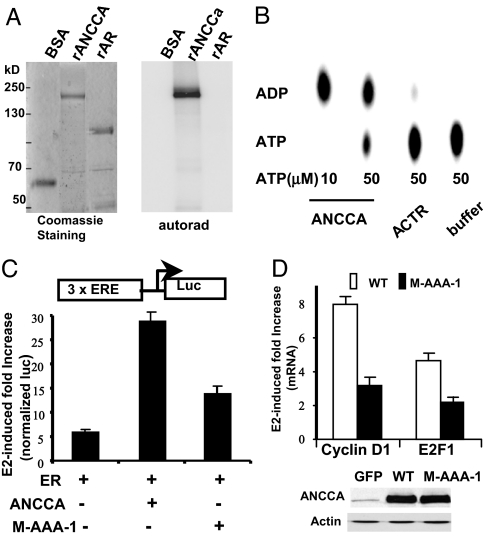
Based on their AAA+ domain sequence homology, AAA+ proteins can be divided into different groups (20). Our sequence analysis indicates that the two AAA+ modules of ANCCA, particularly AAA-D1, share significant homology with that of the so-called classic clade of AAA+ proteins, which include p97/VCP/Cdc48, Hsp104, and NSF. Not only are the five key sequence motifs for ATP binding and/or hydrolysis (Walker A and B, sensors 1 and 2, and arginine finger) and oligomerization (arginine finger) essentially identical to those of p97/VCP, but the sequences between the motifs are also highly conserved (SI Fig. 7B). Therefore, we examined whether ANCCA binds to ATP and possesses ATPase activity using purified recombinant proteins. As expected, full-length ANCCA displayed specific ATP binding activity whereas androgen receptor protein that was expressed and purified similarly showed no detectable ATP binding (Fig. 6A). Consistent with the ATP binding results, we found that ANCCA protein showed strong ATPase activity with an approximate specific activity of 1 μmol/h per milligram, which is comparable to other AAA+ proteins (20). In contrast, ACTR and AR proteins showed no significant ATPase activity (Fig. 6B and data not shown).
Fig. 6.
ANCCA ATPase activity is critical for its function as an ERα coactivator. (A) ANCCA binds ATP. Purified, recombinant ANCCA, or control proteins BSA and purified androgen receptor (rAR), and [γ-32P]ATP were cross-linked by UV irradiation. Treated mixtures were separated by SDS/PAGE. Gels were stained with Coomassie blue or exposed to x-ray film. (B) ANCCA hydrolyzes ATP. Purified, recombinant ANCCA or control ACTR protein was analyzed for ATPase activity as described in Materials and Methods. ATP and its product, ADP, were separated by thin-layer chromatography and imaged by PhosphorImager. (C) A luciferase reporter assay was performed in CV-1 cells with 3× ERE-luc and expression constructs indicated for ERα and ANCCA (wild type or mutant M-AAA-1 with Walker A and Walker motifs mutated in AAA-D1 module). Transfected cells were treated with 10−9 M E2 for 36 h before harvesting for β-gal and luciferase assays. (D) Hormone-deprived cells were infected with adeno-vectors for GFP, wild type, or the mutant ANCCA. Two days after infection, cells were treated with 10−10 M E2 for 12 h before being harvested for real-time RT-PCR analysis or for Western blotting. Fold increase in cyclin D1 and E2F1 mRNA levels was obtained by comparing the quantitative PCR data from E2-treated cells infected with adeno-GFP with the data from E2-treated cells infected with adeno-ANCCA.
The ATPase Is Important for ANCCA to Mediate E2 Stimulation of Gene Expression.
Consistent with the notion that ANCCA is a coactivator of ERα, coexpression of ERα with wild-type ANCCA resulted in a markedly increased E2 response (28-fold in ERα plus ANCCA versus 5-fold in ER alone) of ERE-tk-luciferase reporter activity (Fig. 6C). To eliminate its ATP binding and/or hydrolysis function, we made specific mutations in the Walker A (K473 to T) and Walker B (E532 to Q) motifs of ANCCA AAA-D1 (21, 27). Importantly, in the same reporter gene assay, this mutant form of ANCCA (designated as M-AAA-1) displayed much diminished activity (≈12-fold of E2 response) to enhance the transcriptional activation by ERα. To further examine the importance of AAA-D1 ATPase, we ectopically expressed wild type and the mutant ANCCA in T-47D cells and measured endogenous ERα target gene expression stimulated by a low concentration of E2 (10−10 M). Although wild type and the M-AAA-1 mutant were expressed at a similar level (Fig. 6D Lower), the mutant ANCCA showed significantly reduced ability compared with wild type to enhance the E2 induction of cyclin D1 (3-fold versus 8-fold) and E2F1 (2-fold versus 4.5-fold) expression. Together with the reporter assay data, these results suggest that the ATPase harbored in AAA-D1 of ANCCA is critical for its function as an ERα coactivator to mediate E2 induction of gene expression.
Discussion
We report here our finding of an AAA+ ATPase protein, ANCCA, as a key mediator for E2 signaling and its unique functional mode previously unrecognized for nuclear hormone receptor coactivators. Our results strongly suggest that ANCCA is a bona fide coactivator of ERα with unique features. One of them is its expression induction by E2, which underscores the functional importance of ANCCA in mediating E2 signaling. This positive feedback loop may not only reinforce ERα control of specific gene expression but also expand the E2-responsive gene repertoire as well. Recently, multiple E2-responsive gene networks have been identified (28–33). How these gene networks are coordinated in their expression is unclear. Like ACTR, ANCCA may facilitate the function of both nuclear receptors and nonreceptor transcription factors. The other unique feature is its novel functional mode. Our loss-of-function experiments revealed that ANCCA is important specifically for the recruitment and/or occupancy of CBP at ERα target gene promoters, because its depletion diminishes CBP occupancy at the promoters without affecting that of ERα. This is somewhat unexpected from the docking model of coregulator assembly, because CBP and its related protein, p300, are known to directly interact with nuclear receptors and the p160/SRC coactivators. Therefore, our findings support a new model where one mechanism of ANCCA function is to facilitate the assembly of coregulator complexes at the local chromatin. Based on the virtue of AAA+ protein function, one can speculate that ATP binding and/or hydrolysis in the putative ANCCA oligomer alters ERα and/or ACTR conformation or their configuration in a manner that allows efficient assembly of other protein complexes such as CBP/p300 and the Mediator complex. Consistent with this model, we found that ANCCA displays ATP binding and hydrolysis activities. Moreover, we found that mutation of the Walker A and Walker B motifs that would cripple its ATPase activity diminishes its ability to mediate E2 induction of gene expression. Together, these findings strongly suggest that ANCCA is a novel AAA+ protein that plays a unique function in transcriptional regulation in mammalian cells. Further study will be needed to define the exact mechanism of ANCCA function in coregulator assembly and to determine whether it can also function as a Brg1-type chromatin-remodeling protein, given its ATPase activity and bromodomain, and to examine its relationship to other mechanisms such as posttranslational modifications in modulation of ER coactivator complex assembly (10, 34–37).
ANCCA is also distinct from other ER coactivators such as the p160/SRCs in its selective control of ER target genes. Intriguingly, unlike p160/SRCs, high levels of ANCCA appear not required for the expression of pS2/TFF1 and cathepsin D. Although the latter two have long been characterized as genes highly responsive to E2 stimulation in breast cancer cells, their direct involvement in control of cell proliferation, if any, is unlikely via modulating the cell cycle. On the other hand, the function of ANCCA in mediating E2 induction of cyclin D1, myc, and E2F1 is highly remarkable. These three genes have well established roles in control of cell cycle progression and are often amplified and/or overexpressed in breast cancers (38–42). Therefore, it is tempting to speculate that ANCCA may coordinate the expression of a subset of ER target genes with specific functions such as control of cell cycle. Consistent with the essential role of E2-induced ANCCA in promoting breast cancer cell proliferation, which we demonstrated in this study, recent RNA expression profiling studies using large cohorts of clinical breast cancer tissues identified ANCCA (listed as pro2000 or atad2) among a small number of genes or so-called “gene signatures” that classify ER-positive breast cancers with poor prognosis (43, 44). Our recent immunohistochemistry study suggests that ANCCA protein is overexpressed in a subset of human breast cancers (unpublished data). Therefore, further study of ANCCA function will provide new insights into the mechanism of coregulators in mediating hormone control of gene expression and the potential role of ANCCA in hormonal responsive tumorigenesis.
Materials and Methods
Details.
See SI Materials and Methods for additional information regarding plasmids, viral vectors, RNAi sequences, recombinant proteins, Northern blotting, antibody generation, Western blotting, quantitative RT-PCR, and reporter gene assay.
Cell Proliferation and FACS Analysis of Cell Cycle.
T-47D cells were seeded at 2 × 105 per well in six-well plates and maintained in hormone-depleted medium for 24 h before being infected with equal titers of adeno-RNAi-ANCCA or adeno-RNAi-GFP. Cells were then maintained in hormone-depleted medium for 48 h before being treated with 10 nM E2. MCF-7 cells were transfected with synthetic siRNA as above. Two days after the RNAi treatments, cells were treated with 10 nM E2. Medium was changed every other day, and cell proliferation was measured by cell counting of coded samples in triplicate. For flow cytometry, cells were treated with E2 for 24 h before being detached, fixed in 70% ethanol, stained with propidium iodide, and analyzed for cell cycle distribution by FACScan as previously described (45).
Treatment with siRNA and ChIP Assay.
Cells were plated in six-well plates in regular growth medium and transfected the next day at a density of ≈50% confluence with synthetic siRNA using Dharmafect (Dharmacon) following the manufacturer's protocols. Transfected cells were maintained in hormone-depleted medium for 3 days before being treated with E2 for the indicated times. ChIP assays were performed essentially as described previously (46) with the following modifications. Briefly, ≈2 × 106 cells were fixed with 1% formaldehyde for 8 min at room temperature and lysed in the lysis buffer containing 0.1% SDS. The lysate was then sonicated for 20 min on a Bioruptor (Diagenode). The crude chromatin solutions were incubated overnight at 4°C with the following specific antibodies: anti-ERα (C-20; Santa Cruz Biotechnology, Santa Cruz, CA) at 2 μg/ml, anti-CBP (1:1 mixture of C-20 and A22; Santa Cruz Biotechnology) at 2 μg/ml, anti-acetyl-histone H4 (ChIP grade, 06-866; Upstate Biotechnology) at 5 μl/ml, anti-ACTR (45) at 10 μl/ml, and anti-ANCCA antibody at 2 μg/ml. Precipitated DNA were reverse cross-linked overnight at 65°C, purified, and analyzed as previously described (46).
Coimmunoprecipitation and GST Pull-Down Assay.
MCF-7 cells were treated with 10 nM E2 for 45 min before being harvested for preparation of nuclear extracts. Briefly, cell pellets were resuspended with swelling buffer containing 10 mM Hepes (pH 7.9), 0.75 mM spermidine, 0.15 mM spermine, 0.1 mM EDTA, 0.1 mM EGTA, 10 mM KCl, and 1 mM DTT and incubated on ice for 10 min. After homogenization, the nuclei were collected by brief centrifugation and resuspended in buffer containing 20 mM Hepes (pH 7.9), 0.42 M NaCl, 0.75 mM spermidine, 0.15 mM spermine, 0.2 mM EDTA, 2 mM EGTA, 2 mM DTT, 1 mM PMSF, protease inhibitor mixture (Sigma), and 20% glycerol. The suspension was rocked vigorously at 4°C for 30 min, and the resulting extract was clarified by centrifugation at 30,000 × g for 30 min at 4°C. The recovered supernatant was diluted 1:3 with dilution buffer (20 mM Hepes, pH 7.9/0.2 mM EDTA/1 mM NaF/10 mM Na-pyrophosphate). Diluted nuclear lysates were incubated with equal amounts of anti-ANCCA IgG or control rabbit IgG antibodies for 2 h at 4°C, followed by incubation with protein A beads (Zymed) for 1 h at 4°C. After extensive washing, the immunoprecipitates were analyzed by Western blotting with anti-ER and anti-ACTR monoclonal antibodies (45). For GST pull-down assay, flag-tagged ANCCA protein (≈100 ng) purified from baculovirus-infected Sf9 cells or ANCCA produced in the presence of [35S]methionine were incubated with bead-bound GST fusion or flag-tagged proteins at 4°C for 1 h in a binding buffer containing 20 mM Hepes (pH 7.9), 150 mM NaCl, 1 mM EDTA, 4 mM MgCl2, 1 mM DTT, 0.05% Nonidet P-40, 1 mM PMSF, protease inhibitor mixture (Sigma), 10% glycerol, and 2 mg/ml BSA. For interactions between ANCCA and receptors, 10−7 M E2 or retinoic acid was included in both binding and washing buffer. The beads were washed four times with the binding buffer containing 300 mM KCl and resuspended in SDS/PAGE sample buffer. ANCCA proteins retained on the beads were separated by SDS/PAGE and visualized by Western blotting or autoradiography.
ATP Binding and ATPase Assay.
ANCCA ATP binding was examined by UV cross-linking. Briefly, tubes containing 1 μg of recombinant ANCCA or other proteins in 20 μl of buffer (150 mM NaCl/10 mM MgCl2/1 μCi of [γ-32P]ATP/25 mM Tris·Cl, pH 8.0/0.5 mM PMSF/0.5 mM DTT/7.5% glycerol) were exposed to a germicidal UV lamp for 20 min on ice. Protein samples were then separated by SDS/PAGE and processed for autoradiography. ATP hydrolysis assay was performed as previously described with modifications (47). Briefly, ≈0.5 μg of ANCCA or control proteins was mixed in 15 μl of buffer containing 17 mM Tris·Cl (pH 8.0), 100 mM NaCl, 10 mM MgCl2, 0.3 mM PMSF, 0.3 mM DTT, and 5% glycerol. The reaction was set off by adding 1 μCi of [α-32P]ATP and cold ATP and stopped by addition of 0.5 M EDTA. The reaction mixture was separated on a thin-layer chromatography plate and exposed to a PhosphorImager.
Supplementary Material
Acknowledgments
We thank Dr. Maggie Louie for initial ACTR target gene profiling and Dr. Hsing-Jien Kung for valuable discussion and support. This work was supported by National Institutes of Health Grants DK60019 and CA113860.
Abbreviations
- E2
17β-estradiol
- ER
estrogen receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705814104/DC1.
References
- 1.Hewitt SC, Harrell JC, Korach KS. Annu Rev Physiol. 2005;67:285–308. doi: 10.1146/annurev.physiol.67.040403.115914. [DOI] [PubMed] [Google Scholar]
- 2.Dahlman-Wright K, Cavailles V, Fuqua SA, Jordan VC, Katzenellenbogen JA, Korach KS, Maggi A, Muramatsu M, Parker MG, Gustafsson JA. Pharmacol Rev. 2006;58:773–781. doi: 10.1124/pr.58.4.8. [DOI] [PubMed] [Google Scholar]
- 3.Acevedo ML, Kraus WL. Essays Biochem. 2004;40:73–88. doi: 10.1042/bse0400073. [DOI] [PubMed] [Google Scholar]
- 4.Smith CL, O'Malley BW. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Kinyamu HK, Archer TK. Mol Endocrinol. 2006;20:1–13. doi: 10.1210/me.2005-0192. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG. Mol Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Tremblay GB, Giguere V. Crit Rev Eukaryotic Gene Expression. 2002;12:1–22. doi: 10.1615/critreveukaryotgeneexpr.v12.i1.10. [DOI] [PubMed] [Google Scholar]
- 8.Singh RR, Kumar R. J Cell Biochem. 2005;96:490–505. doi: 10.1002/jcb.20566. [DOI] [PubMed] [Google Scholar]
- 9.Rosenfeld MG, Lunyak VV, Glass CK. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 11.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 12.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 13.Burakov D, Crofts LA, Chang CP, Freedman LP. J Biol Chem. 2002;277:14359–14362. doi: 10.1074/jbc.C200099200. [DOI] [PubMed] [Google Scholar]
- 14.Lee DY, Teyssier C, Strahl BD, Stallcup MR. Endocr Rev. 2005;26:147–170. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- 15.Belandia B, Orford RL, Hurst HC, Parker MG. EMBO J. 2002;21:4094–4103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu W, Cho H, Kadam S, Banayo EM, Anderson S, Yates JR, III, Emerson BM, Evans RM. Genes Dev. 2004;18:144–156. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang ZQ, Li J, Sachs LM, Cole PA, Wong J. EMBO J. 2003;22:2146–2155. doi: 10.1093/emboj/cdg219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acevedo ML, Lee KC, Stender JD, Katzenellenbogen BS, Kraus WL. Mol Cell. 2004;13:725–738. doi: 10.1016/s1097-2765(04)00121-2. [DOI] [PubMed] [Google Scholar]
- 19.Indiani C, O'Donnell M. Nat Rev Mol Cell Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 20.Erzberger JP, Berger JM. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 21.Hanson PI, Whiteheart SW. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 22.Burton BM, Baker TA. Protein Sci. 2005;14:1945–1954. doi: 10.1110/ps.051417505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coser KR, Chesnes J, Hur J, Ray S, Isselbacher KJ, Shioda T. Proc Natl Acad Sci USA. 2003;100:13994–13999. doi: 10.1073/pnas.2235866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngwenya S, Safe S. Endocrinology. 2003;144:1675–1685. doi: 10.1210/en.2002-0009. [DOI] [PubMed] [Google Scholar]
- 26.O'Lone R, Frith MC, Karlsson EK, Hansen U. Mol Endocrinol. 2004;18:1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- 27.Ye Y, Meyer HH, Rapoport TA. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. Proc Natl Acad Sci USA. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stender JD, Frasor J, Komm B, Chang KC, Kraus WL, Katzenellenbogen BS. Mol Endocrinol. 2007;21:2112–2123. doi: 10.1210/me.2006-0474. [DOI] [PubMed] [Google Scholar]
- 30.Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, et al. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 34.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 35.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Proc Natl Acad Sci USA. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park KJ, Krishnan V, O'Malley BW, Yamamoto Y, Gaynor RB. Mol Cell. 2005;18:71–82. doi: 10.1016/j.molcel.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Feng Q, Yi P, Wong J, O'Malley BW. Mol Cell Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butt AJ, McNeil CM, Musgrove EA, Sutherland RL. Endocr Relat Cancer. 2005;12(Suppl 1):S47–S59. doi: 10.1677/erc.1.00993. [DOI] [PubMed] [Google Scholar]
- 39.McNeil CM, Sergio CM, Anderson LR, Inman CK, Eggleton SA, Murphy NC, Millar EK, Crea P, Kench JG, Alles MC, et al. J Steroid Biochem Mol Biol. 2006;102:147–155. doi: 10.1016/j.jsbmb.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 40.Vuaroqueaux V, Urban P, Labuhn M, Delorenzi M, Wirapati P, Benz CC, Flury R, Dieterich H, Spyratos F, Eppenberger U, et al. Breast Cancer Res. 2007;9:R33. doi: 10.1186/bcr1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang SY, Liu SC, Al-Saleem LF, Holloran D, Babb J, Guo X, Klein-Szanto AJ. Cancer Epidemiol Biomarkers Prev. 2000;9:395–401. [PubMed] [Google Scholar]
- 42.Arnold A, Papanikolaou A. J Clin Oncol. 2005;23:4215–4224. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 44.Teschendorff AE, Naderi A, Barbosa-Morais NL, Pinder SE, Ellis IO, Aparicio S, Brenton JD, Caldas C. Genome Biol. 2006;7:R101. doi: 10.1186/gb-2006-7-10-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louie MC, Zou JX, Rabinovich A, Chen HW. Mol Cell Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Louie MC, Revenko AS, Zou JX, Yao J, Chen HW. Mol Cell Biol. 2006;26:3810–3823. doi: 10.1128/MCB.26.10.3810-3823.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zakalskiy A, Hogenauer G, Ishikawa T, Wehrschutz-Sigl E, Wendler F, Teis D, Zisser G, Steven AC, Bergler H. J Biol Chem. 2002;277:26788–26795. doi: 10.1074/jbc.M201515200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.