Abstract
Inwardly rectifying potassium (Kir) channels form gates in the cell membrane that regulate the flow of K+ ions into and out of the cell, thereby influencing the membrane potential and electrical signaling of many cell types, including neurons and cardiomyocytes. Kir-channel function depends on other cellular proteins that aid in the folding of channel subunits, assembly into tetrameric complexes, trafficking of quality-controlled channels to the plasma membrane, and regulation of channel activity at the cell surface. We used the yeast Saccharomyces cerevisiae as a model system to identify proteins necessary for the functional expression of a mammalian Kir channel at the cell surface. A screen of 376 yeast strains, each lacking one nonessential protein localized to the early secretory pathway, identified seven deletion strains in which functional expression of the Kir channel at the plasma membrane was impaired. Six deletions were of genes with known functions in trafficking and lipid biosynthesis (sur4Δ, csg2Δ, erv14Δ, emp24Δ, erv25Δ, and bst1Δ), and one deletion was of an uncharacterized gene (yil039wΔ). We provide genetic and functional evidence that Yil039wp, a conserved, phosphoesterase domain-containing protein, which we named “trafficking of Emp24p/Erv25p-dependent cargo disrupted 1” (Ted1p), acts together with Emp24p/Erv25p in cargo exit from the endoplasmic reticulum (ER). The seven yeast proteins identified in our screen likely impact Kir-channel functional expression at the level of vesicle budding from the ER and/or the local lipid environment at the plasma membrane.
Keywords: yeast deletion library, YIL039W/TED1, GPI anchor, GIRK2, epistasis mini array profile
Inwardly rectifying potassium (Kir) channels serve important physiological functions by regulating the membrane potential of many cell types, including neurons, cardiomyocytes, and hormone-secreting cells. Disruption of Kir-channel function has been linked to human diseases such as periodic paralysis and neonatal diabetes (1).
Kir-channel activity at the plasma membrane is influenced by the abundance of channels and by their functional properties. The number of channels at the cell surface is regulated at the level of channel transcription, biosynthesis, trafficking, and turnover (2). The functional properties of Kir channels are influenced by the membrane potential, local lipid environment, small molecules, and interacting proteins (3, 4). Structure-function studies have identified amino acid motifs and structural features of Kir channels involved in folding, assembly, and trafficking, as well as in gating and selectivity (5–7). However, less is known about the cellular machinery that interacts with these motifs and allows Kir channels to reach the cell surface and function appropriately. We took advantage of the knowledge gained from structure-function studies of Kir channels and the genetic tools available in the yeast Saccharomyces cerevisiae to design a yeast screen aimed at identifying cellular proteins that play a role in Kir-channel functional expression.
We chose to study the mammalian G protein-activated Kir channel GIRK2 (Kir3.2) that can form homotetrameric channels and that mediates inhibitory postsynaptic potentials in midbrain dopamine neurons (8). The mutation S177W (referred to as Kir*) renders Kir3.2 constitutively open in the absence of G protein signaling, permeable to Na+ and K+, and does not disrupt functional expression of the channel at the cell surface of yeast or Xenopus oocytes (9, 10). Expression of mutated K+ channels that are permeable to Na+ overwhelms the Na+ detoxification systems of yeast (11). Functional expression of Kir* can therefore be assayed on the basis of growth inhibition, reflected by small yeast-colony size, on media containing high Na+ concentrations. We reasoned that growth inhibition conferred by Kir* could be overcome if channel biogenesis, trafficking, or function were disrupted.
The Saccharomyces Genome Deletion Project has generated a library of yeast strains each lacking one nonessential gene (12). Additional transgenes can be introduced into the deletion strains by using methods developed for synthetic genetic array (SGA) analysis (13, 14). We used these tools to introduce an inducible Kir* transgene into 376 yeast deletion strains, each lacking an early secretory pathway-localized protein (15), and tested the resulting strains for growth inhibition on high Na+ media conferred by Kir*. We identified seven yeast deletion strains with reduced growth inhibition on high Na+ media, indicating that the strains are missing a gene involved in Kir* functional expression.
Results
Kir* Slows Yeast Growth on High Na+ Media.
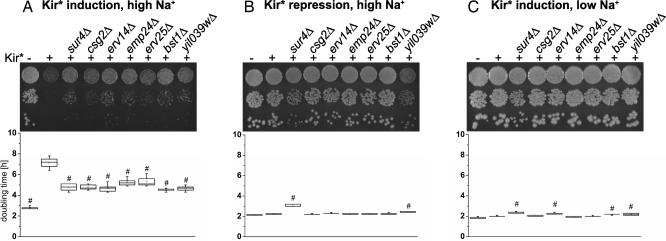
Kir3.2S177W tagged with GFP at the C terminus (Kir*) was integrated into the genome of yeast (BY4742 background) under the control of a galactose-inducible/dextrose-repressible promoter. Whereas yeast not carrying Kir* doubled every 3 h in rich galactose media (YPAGR) media containing 500 mM Na+, expression of Kir* slowed the doubling time to 7 h (Fig. 1A). The inhibition of yeast growth by Kir* was also observed on solid media containing 500 mM Na+ (Fig. 1A). Integration of the channel into the yeast genome did not affect yeast growth when channel expression was repressed by dextrose (Fig. 1B) or under low-sodium conditions (Fig. 1C). Growth on high Na+ media of yeast expressing Kir* was rescued in the vicinity of a filter disk containing the Kir channel blocker barium (16) [supporting information (SI) Fig. 6], supporting the conclusion that growth inhibition conferred by Kir* was due to Na+ influx through the channel.
Fig. 1.
Deletion of seven early secretory pathway-localized proteins reduced Na+ toxicity conferred by Kir*. Growth of yeast strains carrying Kir* alone or in combination with the deletions was assayed by 10-fold serial dilutions on agar plates (Upper) or by doubling time measurements in liquid culture (Lower). (A) Expression of Kir* in control yeast slowed growth in 500 mM Na+ YPAGR. Growth inhibition by Kir* was partially reversed in yeast strains carrying deletions of seven early secretory pathway-localized proteins. (B and C) The deletions did not enhance growth in high-Na+ media when Kir* was repressed (500 mM Na+-rich dextrose media) (B) or in low-Na+ media when Kir* was induced (YPAGR) (C). Whiskers indicate minimum and maximum. Boxes indicate 25th to 75th percentile and median. Open squares indicate the mean, n = 5. #, statistically significant difference compared with yeast expressing Kir* in the control background (P < 0.01, Dunnett's test).
Yeast Screen.
Using the mating and random spore selection scheme developed for SGA analysis (13, 14), we introduced the genomically integrated copy of Kir* into 376 strains from the MATa (BY4741) yeast deletion library (12), each carrying a deletion of an early secretory pathway-localized protein (15) (see SI Table 2 for a list of the deletions and SI Table 3 for the selection scheme). Growth of the deletion strains carrying Kir* was tested on high-Na+ media containing galactose to induce Kir* expression and, to account for strain-specific growth differences, normalized to growth on high-Na+ media containing dextrose, which repressed Kir* expression. Most deletion strains behaved like control (BY4741) yeast and showed growth inhibition on high-Na+ media when Kir* was expressed. However, several strains grew into large colonies even though Kir* expression was induced. Follow-up tests of the Na+-tolerant strains in liquid culture identified seven yeast deletion strains (sur4Δ, csg2Δ, erv14Δ, emp24Δ, erv25Δ, bst1Δ, and yil039wΔ) that grew well under high-Na+, Kir*-inducing conditions.
Deletion Strains Resistant to Growth Inhibition by Kir*.
The candidates fell into two categories (Table 1). The first category was enzymes involved in sphingolipid biosynthesis: Sur4p, which catalyzes the formation of very-long-chain fatty acids (17), and Csg2p, a regulatory subunit of the complex that attaches mannose to inositol phosphorylceramide (18). The second category was proteins involved in cargo selection and vesicle budding during endoplasmic reticulum (ER)-Golgi trafficking: Erv14p, a protein required for packaging of specific cargo into COPII vesicles (19, 20); Emp24p and Erv25p, p24 proteins that form a complex involved in COPII vesicle budding and trafficking of glycosylphosphatidylinositol (GPI)-anchored and soluble proteins (21); Bst1p, an enzyme that removes the acyl chain from GPI anchors, thereby allowing GPI-anchored proteins to leave the ER (22, 23); and Yil039wp, a conserved, metallophosphoesterase domain-containing protein with previously unknown function.
Table 1.
Functions of proteins deleted in strains identified by Kir* screen
| Name | ORF | Localization | Function | Deletion phenotype |
|---|---|---|---|---|
| SUR4 | YLR372W | ER | Elongase for very long-chain fatty acids | Reduced VLCFA levels; lipid raft association and targeting of H+ ATPase disrupted (17, 30, 39, 41) |
| CSG2 | YBR036C | ER | Regulatory subunit of mannosyl-tranferases Csg1p and Csh1p | Reduced mannosylinositol phosphorylceramide levels (18, 40) |
| ERV14 | YGL054C | ER | COPII vesicle packaging chaperone | ER retention of TM proteins Axl2p and Sma2p; delay in ER exit of GPI-anchored proteins (19, 20) |
| EMP24 | YGL200C | COPII vesicles | Cargo receptor in p24 protein family | Delay in ER exit of GPI-anchored proteins and soluble cargo; secretion of ER proteins; suppression of sec13Δ (21, 24, 34–36) |
| ERV25 | YML012W | COPII vesicles | Cargo receptor in p24 protein family | |
| BST1 | YFL025C | ER | GPI inositol deacylase | Delay in ER exit of GPI-AP; secretion of ER proteins; suppression of sec13Δ (22, 23, 36, 48) |
| TED1 | YIL039W | ER | Uncharacterized | Uncharacterized |
To ensure correct identification of the deletions and to rule out differences in the genetic background, mutations in the transgene, or influences of mating type, the seven candidate deletion strains were remade using PCR-mediated gene disruption in the BY4742 background, and the phenotypes were confirmed using growth assays in liquid culture and on agar plates. When Kir* expression was induced by galactose, the seven deletion strains grew faster than the control strain in media containing high Na+ (500 mM YPAGR) (Fig. 1A). The ability of the deletion strains to grow faster in high-Na+ media was not due to general Na+ tolerance, because when Kir* expression was repressed by dextrose, the deletion strains grew at a similar rate or, in the case of sur4Δ and yil039wΔ, more slowly than the control strain in media containing high Na+ (500 mM rich dextrose media) (Fig. 1B). The deletions did not enhance the ability of the yeast to metabolize galactose, as shown by comparable or slower growth in galactose containing media under conditions of low Na+ (YPAGR, ≈30 mM Na+) (Fig. 1C). Finally, Na+ tolerance under Kir*-inducing conditions was not explained by osmotolerance, because the deletion strains grew at similar rates or more slowly than control yeast in hyperosmotic media containing 1 M sorbitol (data not shown).
Although the deletion strains expressing Kir* grew faster than the control strain expressing Kir* in 500 mM Na+ YPAGR (Fig. 1A), they did not grow as fast as a control strain without genomic insertion of Kir*, likely because the deletions did not entirely abolish Kir* function at the plasma membrane. Such partial rescue would be expected for deletions affecting trafficking or quality control, which often employ backup pathways (24, 25). In addition, the Na+ sensitivity (Fig. 1B) and slow growth in galactose media (Fig. 1C) of some of the strains (sur4Δ, erv14Δ, bst1Δ, yil039WΔ) may have contributed to the incomplete rescue, because for these strains even complete loss of the Kir* function would not have resulted in the same growth as control yeast not carrying Kir*.
On the basis of the result that reduced growth inhibition of the deletion strains depended on Kir* expression in the presence of high Na+, we concluded that Kir* functional expression at the plasma membrane was disrupted in these strains. However, it was also possible that the membrane potential of the deletion strains was depolarized.
Hygromycin B Sensitivity of Deletion Strains.
Na+ influx through Kir* is driven by the hyperpolarized membrane potential of yeast. Therefore, growth inhibition by high Na+ would be reduced if the deletion strains had more depolarized membrane potentials than control yeast. The small size of yeast precludes electrophysiological measurements of their membrane potential; however, relative membrane potentials can be assayed based on uptake of lipophilic cations or sensitivity to the antibiotic hygromycin B (26–28). We therefore tested whether the seven deletion strains were hygromycin-resistant, indicative of depolarization, compared with control yeast. To ensure that our assay would detect depolarization of the membrane potential, we tested the yeast strain pma1–105, which carries a mutation in the proton ATPase Pma1p and has previously been shown to be depolarized (28, 29). Growth of the pma1–105 strain was inhibited by hygromycin B less than was growth of the corresponding control DBY745 strain (Fig. 2A). Comparing the deletion strains identified in our screen with the corresponding control BY4742 strain (Fig. 2B), the sur4Δ and erv14Δ strains were slightly less inhibited by hygromycin, indicating that they may be more depolarized. Hygromycin resistance has been reported for sur4-mutant strains in the BWG1–7A genetic background (30). However, the differences in relative growth rates in our experiment were not statistically significant (Dunnett's test comparing BY4742 with each deletion strain, P > 0.05). Because hygromycin B sensitivity cannot be calibrated in terms of absolute changes in membrane potential, we cannot rule out that the tendency toward hygromycin resistance in sur4Δ and erv14Δ strains accounted, at least in part, for the reduced growth inhibition by Na+ influx through Kir*. The csg2Δ strain showed a tendency (not significant by Dunnett's test, P > 0.05) toward increased hygromycin sensitivity, and the emp24Δ, erv25Δ, bst1Δ, and yil039wΔ strains had comparable hygromycin sensitivity to the control strain, suggesting that depolarization did not account for the ability of these deletion strains to grow under high-Na+ conditions while expressing Kir*.
Fig. 2.
Hygromycin B sensitivity of deletion strains. Growth rates measured in 500 mM Na+ YPAGR liquid media with 500 mg/liter hygromycin B were normalized to growth rates in 500 mM Na+ YPAGR. (A) The assay detected hygromycin resistance of pma1–105 yeast compared with control DBY745 yeast (P < 0.01, t test). (B) The seven deletion strains showed no significant difference in hygromycin sensitivity compared with control BY4742 yeast (P > 0.05, Dunnett's test), although the csg2Δ strain showed a tendency toward increased hygromycin sensitivity, and the sur4Δ and erv14Δ strains showed a tendency toward increased hygromycin resistance. Error bars are standard errors, n = 3.
Impaired Complementation of trk1Δ trk2Δ Yeast by Kir3.2V188G.
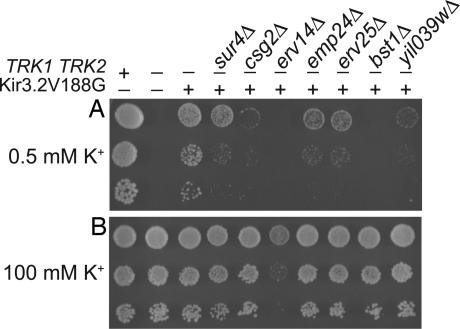
To corroborate that the seven deletions impaired functional expression of Kir* at the cell surface, we used an independent assay. Yeast lacking the K+ transporters Trk1p and Trk2p are starved for K+ and therefore grow slowly on low-salt media supplemented with low concentrations (0.5 mM) of K+ (31). Growth is rescued by expression of Kir3.2V188G, a constitutively active, K+-selective Kir3.2 channel (9). If the deletions identified in our screen disrupted Kir-channel trafficking or function, we predicted that rescue of trk1Δ trk2Δ yeast by Kir3.2V188G would be impaired in the deletion background. Indeed, Kir3.2V188G rescued growth on 0.5 mM K+ media poorly or not at all when trk1Δ trk2Δ yeast carried a deletion of SUR4, CSG2, EMP24, ERV25, BST1, or YIL039W (Fig. 3). These yeast strains grew well on low-salt media supplemented with 100 mM K+, on which they did not depend on functional expression of Kir3.2V188G. The erv14Δ trk1Δ trk2Δ strain expressing Kir3.2V188G could not be tested in this assay, because the strain grew slowly on low-salt plates even when supplemented with 100 mM K+.
Fig. 3.
The seven deletions impaired rescue of trk1Δ trk2Δ yeast by Kir3.2V188G. Ten-fold serial dilutions were spotted onto low-salt plates containing 0.5 mM KCl or 100 mM KCl. (A) trk1Δ trk2Δ yeast did not grow on 0.5 mM K+ media. Growth was rescued by expression of Kir3.2V188G. In triple-mutant yeast lacking Trk1p, Trk2p, and one of seven early secretory pathway-localized proteins, Kir3.2V188G only partially restored growth on 0.5 mM K+ media. (B) The triple mutant yeast strains, except erv14Δ, grew well on 100 mM K+ media, in which Kir3.2V188G is dispensable for growth.
Kir* Expression Levels and Localization in the Deletion Strains.
The Na+-tolerant phenotype, impaired rescue of trk1Δ trk2Δ yeast, and the known functions of Sur4p, Csg2p, Erv14p, Emp24p, Erv25p, and Bst1p, suggested that the deletions might have affected Kir-channel maturation or trafficking. We therefore performed Western blot analysis on each of the strains to test whether the deletions altered the total protein levels of Kir*. Similar amounts of Kir* were present in samples from yeast expressing Kir* in the control or deletion background (Fig. 4A).
Fig. 4.
Total protein levels and distribution of Kir*-GFP. (A) Western blot analysis of yeast expressing Kir* in the control or deletion background was probed with anti-Kir3.2 antibody. A band of similar intensity was detected in all strains carrying Kir*. Phosphoglycerate kinase (PGK) served as a loading control. Molecular mass markers are 100 and 75 kDa for anti-Kir3.2 and 50 and 37 kDa for anti-PGK. (B) Deconvolved optical z sections through the middle (Left) or periphery (Right) of yeast expressing Kir* tagged with eGFP at the C terminus. In all strains, Kir* localized to the perinuclear and peripheral ER. (Scale bar, 2.5 μm.)
Given comparable expression levels of Kir* in the deletion strains, we examined whether the deletions altered the subcellular localization of the channel. Yeast were grown in galactose-containing media to induce Kir* expression, fixed, and mounted for imaging of the GFP-tagged Kir*. Optical sections through the middle of yeast cells showed two rings of GFP fluorescence, and sections through the periphery of the cells showed tubular distribution of the GFP-tagged channel (Fig. 4B). The pattern of Kir*-GFP fluorescence was typical of ER-localized proteins (32) even in the control strain. This finding was consistent with studies showing heavy ER localization of Kir3.2 in mammalian cells (33). Given the predominant ER localization of Kir* even in the control background, alterations in ER retention in the deletion strains could not be readily detected.
Deletion of YIL039W Slows Gas1p Trafficking.
Six of the seven mutants identified by our screen had well characterized functions impacting trafficking and lipid biosynthesis, which could explain their effects on Kir* channel functional expression (see Discussion). However, it was unclear how the uncharacterized but conserved Yil039wp influenced Kir* activity. A previously published quantitative genetic interaction map suggested that Yil039wp acts together with Emp24p and Erv25p in mediating trafficking of cargo out of the ER (15). In this epistasis mini array profile (E-MAP), colony sizes for all double mutant combinations were used to assess genetic interactions among ≈400 strains each carrying a deletion in an early secretory pathway gene. When strains were clustered based on the similarity in their patterns of genetic interactions, the emp24Δ and erv25Δ strains alongside erp1Δ were most similar to each other out of all 400 strains. This similarity was expected, because Emp24p, Erv25p, and Erp1p act together in a physical complex (34, 35). The next most similar and therefore most functionally related gene was YIL039W. Moreover, the double mutants of yil039wΔ and emp24Δ or erv25Δ displayed buffering genetic interactions (Fig. 5A adapted from ref. 15), i.e., in the absence of Emp24p/Erv25p there was little additional fitness cost to losing Yil039wp. Buffering genetic interactions were also observed using a fluorescent reporter of unfolded protein response (UPR)-induction. Both yil039wΔ and erv25Δ yeast (emp24Δ was not assayed for technical reasons) showed UPR activation. Deletion of YIL039W and ERV25 together did not exacerbate the phenotype to the extent expected for functionally unrelated genes (e.g., ALG3, OST3, and SPC2; Fig. 5B). These relationships indicate that Yil039wp functions in a concerted manner with Emp24p/Erv25p.
Fig. 5.
Ted1p, encoded by YIL039W, is involved in trafficking of the GPI-anchored protein Gas1p. (A) YIL039W, EMP24, and ERV25 were predicted to act in a concerted manner on the basis of their buffering genetic interactions as determined in ref. 15. (B) UPR induction assayed by expression of GFP from a UPR-inducible promoter. Combining deletions of YIL039W and ERV25 did not enhance UPR activation to the extent expected for unrelated genes (e.g., ALG3, OST3, SPC2), suggesting that Yil039wp and Erv25p share a common function. (C) Western blot analysis of whole-cell extracts probed with an antibody to Gas1p. Deletion of YIL039W/TED1 led to accumulation of Gas1p in its 100-kDa core glycosylated ER form, as previously observed for emp24Δ and erv25Δ strains (34, 36).
To directly test whether Yil039wp, Emp24p, and Erv25p share a common function, we investigated whether ER exit of the GPI-anchored protein Gas1p, which is delayed in emp24Δ and erv25Δ strains (34, 36), was affected in the yil039wΔ strain. Western blot analysis of whole-cell extracts showed that Gas1p accumulated in its 100-kDa core-glycosylated ER form to a similar extent in yeast lacking EMP24, ERV25, or YIL039W (Fig. 5C). We therefore named YIL039W “trafficking of Emp24p/Erv25p-dependent cargo disrupted 1” (TED1).
Discussion
Yeast has been used extensively as a model system to study K+-channel structure–function relationships because of its sensitivity to even small currents and easy manipulation, which allows for screening of thousands of mutated channels (11). We chose to study yeast as a model system because of its powerful genetic tools. Because cellular trafficking is a highly conserved process, we reasoned that secretory pathway conditions that influence a mammalian Kir channel in yeast would inform us of similar requirements in less genetically amenable mammalian systems. Taking advantage of the yeast deletion library (12) and SGA methodology (13, 14), we found that deletion of SUR4, CSG2, ERV14, EMP24, ERV25, BST1, or YIL039W/TED1 impaired Kir-channel functional expression: First, the deletions partially restored yeast growth on high-Na+ media in the presence of the mutated, Na+-permeable K+ channel Kir3.2S177W (Kir*). Second, a K+-selective Kir channel (Kir3.2V188G) was unable to rescue growth on low-K+ media of trk1Δ trk2Δ yeast also carrying one of the deletions.
A common theme among five of the proteins identified by our screen (Erv14p, Emp24p, Erv25p, Bst1p, and Yil039wp/Ted1p) is that they affect maturation and trafficking of GPI-anchored proteins. This finding was unexpected, because Kir channels are transmembrane proteins not known to be modified by a GPI anchor. It is possible that the machinery required for ER exit of GPI-anchored proteins has additional functions in trafficking of transmembrane proteins. In fact, deletion of Erv14p leads to ER retention of the transmembrane proteins Axl2p and Sma1p (19, 20). Alternatively, GPI-anchored proteins may indirectly affect Kir-channel trafficking. Slowed ER exit of GPI-anchored proteins in gwt1–10 yeast has been shown to disrupt the formation of lipid domains in the ER and thereby to indirectly affect sorting and budding of transmembrane proteins (37). We speculate that the interplay between different classes of proteins during the formation of lipid microdomains (38) may affect trafficking of Kir channels.
Deletion of the other two candidates identified by our screen, SUR4 or CSG2, alters the lipid composition of yeast cells by reducing synthesis of C24 and C26 fatty acids (17, 39) or of sphingolipids with mannose modification of their headgroups (40), respectively. The lipid composition of membranes may influence Kir-channel functional expression in two ways. First, lipid rafts rich in sphingolipids or their precursor, ceramide, play a role in trafficking at the level of ER exit (41–43) and at the level of protein sorting at the Golgi (44). Second, the local lipid environment at the plasma membrane may influence channel activity. For example, enrichment of membranes with cholesterol induced an inactive channel conformation in Kir2.1 (45), and a specific interaction between the bacterial K+ channel KcsA and phosphatidylglycerol is required for channel function (46). C24 and C26 fatty acids are also found in remodeled GPI anchors (47), opening the possibility that deletion of SUR4 affects Kir channel trafficking through indirect effects on GPI-anchored proteins, as discussed above.
Our screen identified a phenotype for the previously uncharacterized gene YIL039W, which encodes a metallophosphoesterase domain-containing protein conserved in eukaryotes, including humans (MPPE1). Genetic interaction data based on yeast growth (15) and UPR activation, as well as biochemical data showing ER retention of Gas1p in emp24Δ, erv25Δ (34, 36), and yil039wΔ yeast provide evidence that Yil039wp acts together with Emp24p and Erv25p in cargo exit from the ER. We therefore named YIL039W trafficking of Emp24p/Erv25p-dependent cargo disrupted 1 (TED1). It is interesting to note that the bst1Δ strain, in which Gas1p maturation was also delayed (as reported in ref. 48), displayed an aggravating genetic interaction with ted1Δ but buffering interactions with emp24Δ and erv25Δ. We therefore predict that Bst1p and Ted1p function in parallel pathways to regulate Emp24p/Erv25p function. Because Yil039wp/Ted1p contains a predicted phosphoesterase domain, it will be of interest to identify the protein and/or lipid targets that are dephosphorylated by Ted1p. One candidate substrate is the amphiphysin homologue Rvs167p, which is phosphorylated by Pho85-Pcl1 (49) and was shown in a large-scale pull-down study to physically interact with Ted1p (50).§§
Because Kir3.2 is not native to yeast, our screen was intended to identify global requirements for Kir-channel functional expression and probably precluded the identification of specific chaperoning interactions, which would require coevolution. The seven proteins identified by our screen and their cellular roles are conserved up to mammals, highlighting the appropriate nature of yeast as a model system to uncover basic cellular machinery involved in Kir-channel functional expression. The results provide important leads that will allow us to probe deeper into the mechanisms that regulate trafficking and activity of Kir channels in mammalian systems.
Materials and Methods
Yeast Strains and Media.
Yeast strains were picked from the deletion library (12) or constructed by PCR-mediated gene disruption in the BY4742 background (52). SI Table 4 lists strains, primers, and plasmids. Yeast media recipes were based on refs. 11 and 14 or are provided in SI Methods.
Yeast Screen.
Three hundred seventy-six yeast strains from the MATa deletion library (SI Table 2) (12, 15) were mated to yeast expressing Kir3.2S177W-GFP by using SGA methodology (13, 14). The selection scheme is shown in SI Table 3. Growth of the double-mutant strains was tested on synthetic media containing 750 mM Na+ and dextrose or galactose. Plates were photographed using a ChemiImager Ready (Alpha Innotech, San Leandro, CA), and colony sizes, Sgal and Sdex, were measured using software developed in ref. 53. Initial Na+-tolerant candidates had to meet the criterion that four of six replicates or the average of the six colony-size differences |Sgal × 100/Sdex − 100| were smaller than the average |Sgal × 100/Sdex − 100| for all strains tested minus one standard deviation.
Yeast Assays.
Doubling times and growth rates were determined at 30°C by diluting saturated cultures to 2 × 106 cells per milliliter and by measuring the OD660 at 0 h and 4 or 8 h later. For growth tests on plates, over night, liquid cultures were adjusted to equal ODs, and 10-fold serial dilutions were plated. Photographs were taken 3 days after plating. The experiments were repeated at least two times. Yeast protein samples were prepared by the postalkaline lysis method (54). Western blots were probed with rabbit anti-GIRK2 (Alomone, Jerusalem, Israel), mouse anti-PGK (Invitrogen, Carlsbad, CA), or rabbit anti-Gas1p (provided by P.W.) antibodies. Fixed yeast cells were imaged using wide-field epifluorescence on a Nikon (Tokyo, Japan) TE2000 microscope. Images presented are single planes from the middle and top of deconvolved stacks. For the UPR assay, fluorescence signals from 4xUPRE-GFP normalized to TEF2pr-RFP were measured using Flow Cytometry. For detailed procedures see SI Methods.
Statistics.
One-way ANOVA followed by Dunnett's test and unpaired t test were performed with GraphPad (San Diego, CA) Prism 4.0.
Supplementary Material
Acknowledgments
We thank B. Schwappach (University of Manchester, Manchester, U.K.) for the pYES2 plasmid; C. Boone (University of Toronto, Toronto Canada) and A. Tong (University of Toronto) for yeast strains; J. Haber (Brandeis University, Waltham, MA) for the pma1–105 and control strains; S. Collins for the colony measuring software and help with data analysis; R. Shaw for help with image acquisition and use of the microscope; and members of the L.Y.J. and J.S.W. laboratories for stimulating discussions. This work was supported by National Institute of Mental Health MERIT Award R37MH065334. M.S. was supported by the International Human Frontier Science Program Organization and National Institutes of Health Award K99/R00. M.J. was supported by the National Science Foundation. P.W., J.S.W., Y.-N.J., and L.Y.J. are investigators of the Howard Hughes Medical Institute.
Abbreviations
- ER
endoplasmic reticulum
- GPI
glycosylphosphatidylinositol
- Kir
inwardly rectifying K+
- Kir*
Kir3.2S177W
- Kir3.2
the G protein-activated Kir channel GIRK2
- SGA
synthetic genetic array
- UPR
unfolded protein response
- YPAGR
rich galactose media.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708765104/DC1.
Intriguingly, SUR4 was identified as a suppressor of rvs161 and rvs167 (51).
References
- 1.Neusch C, Weishaupt JH, Bahr M. Cell Tissue Res. 2003;311:131–138. doi: 10.1007/s00441-002-0669-x. [DOI] [PubMed] [Google Scholar]
- 2.Deutsch C. Annu Rev Physiol. 2002;64:19–46. doi: 10.1146/annurev.physiol.64.081501.155934. [DOI] [PubMed] [Google Scholar]
- 3.Ruppersberg JP. Pflügers Arch. 2000;441:1–11. doi: 10.1007/s004240000380. [DOI] [PubMed] [Google Scholar]
- 4.Logothetis DE, Jin T, Lupyan D, Rosenhouse-Dantsker A. Pflügers Arch. 2007;455:83–95. doi: 10.1007/s00424-007-0276-5. [DOI] [PubMed] [Google Scholar]
- 5.Tinker A, Jan LY. Curr Topics Membr. 1999;46:143–158. [Google Scholar]
- 6.Ma D, Jan LY. Curr Opin Neurobiol. 2002;12:287–292. doi: 10.1016/s0959-4388(02)00319-7. [DOI] [PubMed] [Google Scholar]
- 7.Bichet D, Haass FA, Jan LY. Nat Rev Neurosci. 2003;4:957–967. doi: 10.1038/nrn1244. [DOI] [PubMed] [Google Scholar]
- 8.Mark MD, Herlitze S. Eur J Biochem. 2000;267:5830–5836. doi: 10.1046/j.1432-1327.2000.01670.x. [DOI] [PubMed] [Google Scholar]
- 9.Yi BA, Lin YF, Jan YN, Jan LY. Neuron. 2001;29:657–667. doi: 10.1016/s0896-6273(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 10.Bichet D, Lin YF, Ibarra CA, Huang CS, Yi BA, Jan YN, Jan LY. Proc Natl Acad Sci USA. 2004;101:4441–4446. doi: 10.1073/pnas.0401195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura RL, Gaber RF. Methods Enzymol. 1998;293:89–104. doi: 10.1016/s0076-6879(98)93009-9. [DOI] [PubMed] [Google Scholar]
- 12.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 13.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 14.Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Methods. 2006;40:344–352. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 15.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 16.Kubo Y, Adelman JP, Clapham DE, Jan LY, Karschin A, Kurachi Y, Lazdunski M, Nichols CG, Seino S, Vandenberg CA. Pharmacol Rev. 2005;57:509–526. doi: 10.1124/pr.57.4.11. [DOI] [PubMed] [Google Scholar]
- 17.Rossler H, Rieck C, Delong T, Hoja U, Schweizer E. Mol Genet Genomics. 2003;269:290–298. doi: 10.1007/s00438-003-0836-0. [DOI] [PubMed] [Google Scholar]
- 18.Uemura S, Kihara A, Iwaki S, Inokuchi J, Igarashi Y. J Biol Chem. 2007;282:8613–8621. doi: 10.1074/jbc.M606649200. [DOI] [PubMed] [Google Scholar]
- 19.Powers J, Barlowe C. J Cell Biol. 1998;142:1209–1222. doi: 10.1083/jcb.142.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakanishi H, Suda Y, Neiman AM. J Cell Sci. 2007;120:908–916. doi: 10.1242/jcs.03405. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser C. Proc Natl Acad Sci USA. 2000;97:3783–3785. doi: 10.1073/pnas.97.8.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka S, Maeda Y, Tashima Y, Kinoshita T. J Biol Chem. 2004;279:14256–14263. doi: 10.1074/jbc.M313755200. [DOI] [PubMed] [Google Scholar]
- 23.Fujita M, Yoko OT, Jigami Y. Mol Biol Cell. 2006;17:834–850. doi: 10.1091/mbc.E05-05-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Springer S, Chen E, Duden R, Marzioch M, Rowley A, Hamamoto S, Merchant S, Schekman R. Proc Natl Acad Sci USA. 2000;97:4034–4039. doi: 10.1073/pnas.070044097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olkkonen VM, Ikonen E. J Cell Sci. 2006;119:5031–5045. doi: 10.1242/jcs.03303. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Navarro A. Biochim Biophys Acta. 2000;1469:1–30. doi: 10.1016/s0304-4157(99)00013-1. [DOI] [PubMed] [Google Scholar]
- 27.Vallejo CG, Serrano R. Yeast. 1989;5:307–319. doi: 10.1002/yea.320050411. [DOI] [PubMed] [Google Scholar]
- 28.Perlin DS, Brown CL, Haber JE. J Biol Chem. 1988;263:18118–18122. [PubMed] [Google Scholar]
- 29.McCusker JH, Perlin DS, Haber JE. Mol Cell Biol. 1987;7:4082–4088. doi: 10.1128/mcb.7.11.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Arranz M, Maldonado AM, Mazon MJ, Portillo F. J Biol Chem. 1994;269:18076–18082. [PubMed] [Google Scholar]
- 31.Ko CH, Buckley AM, Gaber RF. Genetics. 1990;125:305–312. doi: 10.1093/genetics/125.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 33.Ma D, Zerangue N, Raab-Graham K, Fried SR, Jan YN, Jan LY. Neuron. 2002;33:715–729. doi: 10.1016/s0896-6273(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 34.Belden WJ, Barlowe C. J Biol Chem. 1996;271:26939–26946. doi: 10.1074/jbc.271.43.26939. [DOI] [PubMed] [Google Scholar]
- 35.Marzioch M, Henthorn DC, Herrmann JM, Wilson R, Thomas DY, Bergeron JJ, Solari RC, Rowley A. Mol Biol Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elrod-Erickson MJ, Kaiser CA. Mol Biol Cell. 1996;7:1043–1058. doi: 10.1091/mbc.7.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okamoto M, Yoko-o T, Umemura M, Nakayama K, Jigami Y. J Biol Chem. 2006;281:4013–4023. doi: 10.1074/jbc.M504684200. [DOI] [PubMed] [Google Scholar]
- 38.Hancock JF. Nat Rev Mol Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh CS, Toke DA, Mandala S, Martin CE. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 40.Uemura S, Kihara A, Inokuchi J, Igarashi Y. J Biol Chem. 2003;278:45049–45055. doi: 10.1074/jbc.M305498200. [DOI] [PubMed] [Google Scholar]
- 41.Toulmay A, Schneiter R. Biochimie. 2007;89:249–254. doi: 10.1016/j.biochi.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 42.Horvath A, Sutterlin C, Manning-Krieg U, Movva NR, Riezman H. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dupre S, Haguenauer-Tsapis R. Traffic. 2003;4:83–96. doi: 10.1034/j.1600-0854.2003.40204.x. [DOI] [PubMed] [Google Scholar]
- 44.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 45.Romanenko VG, Fang Y, Byfield F, Travis AJ, Vandenberg CA, Rothblat GH, Levitan I. Biophys J. 2004;87:3850–3861. doi: 10.1529/biophysj.104.043273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valiyaveetil FI, Zhou Y, MacKinnon R. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 47.Pittet M, Conzelmann A. Biochim Biophys Acta. 2007;1771:405–420. doi: 10.1016/j.bbalip.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 48.Vashist S, Kim W, Belden WJ, Spear ED, Barlowe C, Ng DT. J Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dephoure N, Howson RW, Blethrow JD, Shokat KM, O'Shea EK. Proc Natl Acad Sci USA. 2005;102:17940–17945. doi: 10.1073/pnas.0509080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 51.Desfarges L, Durrens P, Juguelin H, Cassagne C, Bonneu M, Aigle M. Yeast. 1993;9:267–277. doi: 10.1002/yea.320090306. [DOI] [PubMed] [Google Scholar]
- 52.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 53.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kushnirov VV. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.