Abstract
Activator of G protein signaling 3 (AGS3), originally identified in a functional screen for mammalian proteins that activate heterotrimeric G protein signaling, is known to be involved in drug-seeking behavior and is up-regulated during cocaine withdrawal in animal models. These observations indicate a potential role for AGS3 in the formation or maintenance of neural plasticity. We have found that the overexpression of AGS3 alters the surface-to-total ratios of a subset of heterologously expressed plasma membrane receptors and channels. Further analysis of the endocytic trafficking of one such protein by a biotin-based internalization assay suggests that overexpression of AGS3 moderately affects the internalization or recycling of surface proteins. Moreover, AGS3 overexpression and siRNA-mediated knockdown of AGS3 both result in the dispersal of two endogenously expressed trans-Golgi network (TGN)-associated cargo proteins without influencing those in the cis- or medial-Golgi compartments. Finally, adding a TGN-localization signal to a CD4-derived reporter renders the trafficking of fusion protein sensitive to AGS3. Taken together, our data support a model wherein AGS3 modulates the protein trafficking along the TGN/plasma membrane/endosome loop.
Keywords: drug addiction, Golgi apparatus, membrane trafficking, receptors and channels
Activator of G protein signaling 3 (AGS3) was originally identified during a functional screen for mammalian proteins that activate heterotrimeric G protein signaling in a receptor-independent manner in Saccharomyces cerevisiae (1). Sequence analysis indicates that AGS3 consists of a three-module structure. The N-terminal part of AGS3 contains seven tetratricopeptide repeats [the TPR domain (2)], a mediator of protein–protein interaction, whereas the C-terminal part contains four G protein regulatory motifs [the GPR or GoLoco domain (3)], a modulator of G protein signaling. The GPR domain of AGS3 preferentially binds and stabilizes GDP-bound Gαi subunits (4–6). By acting as a GDP-dissociation inhibitor of the Gαi subunit, AGS3 blocks the reassociation of Gαi with the Gβγ dimer, thus it inhibits the Gαi-dependent pathways but enhances the Gβγ-regulated signaling in a manner independent of receptor activation. Although AGS3 was initially described in the brain and testis, subsequent studies have confirmed its presence in multiple tissue and cell types (1, 5, 7–9). In the heart, two short forms of AGS3 lacking the TPR domain are detected in addition to the full-length AGS3 (7).
There is evidence that AGS3 participates in diverse cellular events, including macroautophagy in human intestinal HT-29 cells (9) and Gβγ-mediated mitotic spindle orientation in cell division of cerebral cortical progenitors (10). In addition, in an animal model of cocaine withdrawal, AGS3 is up-regulated in the prefrontal cortex and the nucleus accumbens, two brain regions essential for the reinstatement of drug-seeking behavior (11, 12). Importantly, knockdown of AGS3 expression by infusing an AGS3 antisense RNA into the prefrontal cortex abolishes the reoccurrence of cocaine-seeking behavior (11). When the infusion is discontinued, this behavior is restored (11). In a separate study, a similar antisense approach used in the nucleus accumbens prevents the relapse of the heroin-seeking phenotype (12). These observations establish a critical role of AGS3 in drug addiction and further imply a potential function of AGS3 in the formation or maintenance of neural plasticity.
Regulation of trafficking of receptors and channels represents one important mechanism in the modulation of neural plasticity. Whereas the involvement of AGS3 in membrane trafficking has not been documented, several lines of evidence are consistent with this hypothesis. First, although subcellular fractionation studies suggest that the majority of AGS3 exists in the cytosolic fraction, a small amount can be found in the particulate form (5, 13). Indeed, AGS3 has been reported to display a partial colocalization with markers of the endoplasmic reticulum (ER) and the Golgi apparatus (8, 14). Second, one major interacting partner of AGS3, Gαi3, is localized primarily at the Golgi apparatus (15, 16). Third, the mammalian homolog of Drosophila melanogaster partner of inscuteable (mPins), a protein closely related to AGS3, was recently shown to interact with two members of the PDZ-domain containing protein family, PSD-95 and SAP102, and promotes the surface expression of NMDA receptors in neurons (17). Based on these observations, we examined whether AGS3 functions in membrane trafficking.
Results
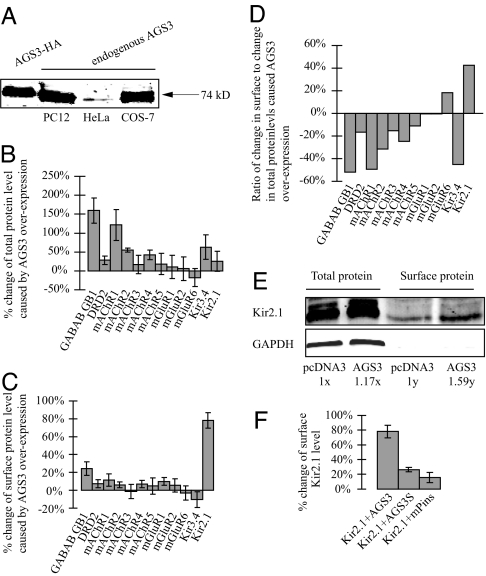
Although previous studies have shown AGS3 expression in a wide variety of cell types, it has not been studied in COS7 or HeLa cells to our knowledge. Using a commercially available antibody, we performed a Western blot and found that both cell types express endogenous AGS3 (Fig. 1A).
Fig. 1.
The effects of overexpressing AGS3 or mPins on the surface and total protein levels of receptors and channels. (A) An antibody against AGS3 (CalBiochem) recognized a 74-kDa band from HeLa and COS7 lysates. Lysates from COS7 cells overexpressing an HA-tagged AGS3 and PC12 cells were included as positive controls (14). (B–F) Extracellular HA-tagged receptors or channels were cotransfected into COS7 cells with an equal amount of empty pcDNA3 vector (as a control) or pcDNA3 expressing AGS3. (B) Total protein levels were determined by a quantitative Western blot analysis using the Li-Cor Odyssey Infrared Imaging System. (C) Surface proteins levels were determined by the previously described chemiluminescence assay (19). (D) The surface-to-total ratio of Kir2.1 is increased by ≈42% in cells overexpressing AGS3. (E) A similar increase (≈36%) of the surface-to-total ratio of Kir2.1 was seen with a complementary biotinylation approach. 1x and 1y are arbitrary units and represent the total and surface levels of control samples, respectively. GAPDH was probed to assure the plasma membrane integrity during biotinylation. (F) Unlike AGS3, AGS3S and mPins both fail to significantly enhance the surface expression of Kir2.1. The biotinylation experiments were repeated twice and similar results were obtained. The values shown in the other graphs are the average of at least six independent experiments.
Overexpression of AGS3 Changes the Ratio of Surface-to-Total Protein Levels of a Subset of Receptors and Channels.
Given the potential link between AGS3 and neural plasticity (11, 12), and the fact that a closely related protein, mPins, facilitates the surface expression of NMDA receptors (17), we decided to assess the impact of overexpressing AGS3 on the surface levels of various G protein-coupled receptors (GPCRs) and inwardly rectifying potassium (Kir) channels in transfected COS7 cells. Each of the receptors and channels was cloned into the expression vector pcDNA3 (Invitrogen, Carlsbad, CA) with identical flanking untranslated regions. One GPCR, the GB1 subunit of GABAB, contained an inactivated ER retention/retrieval signal to facilitate its surface detection (18). All of the GPCRs and Kir channels used in our study contain an extracellular HA epitope to allow quantitative measurement of their surface [using a previously described chemiluminescence assay (19)] or total expression levels [using the Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE)] (Fig. 1 B–F). The HA epitope was inserted at the N termini of GPCRs or in the first extracellular loop of the Kir channels. Previous studies have shown that an epitope insertion in these positions has no detectable effects on the function of Kir channels or GPCRs (19–22). Compared with an empty vector control, overexpression of AGS3 caused an increase in the total protein levels of most receptors and channels assayed (Fig. 1B). However, a similar treatment had little effect on their surface levels, except in the case of Kir2.1 (Fig. 1C). As a result, whereas AGS3 overexpression leads to a decrease or little change of the surface-to-total ratios of most receptors and channels examined, it specifically enhances that of Kir2.1 by 42% (Fig. 1D). A similar increase (≈36%) in the surface-to-total ratio of Kir2.1 was observed when surface biotinylation was used as an alternative approach to quantify the surface pool of channel proteins (Fig. 1E). It has been our observation that a large fraction of expressed receptor and channel proteins reside at the ER and Golgi compartments (data not shown), suggesting that their export from these compartments may be rate-limiting under our experimental conditions. Therefore, we do not know to what extent the AGS3-mediated enhancement of protein level might saturate the transport machinery leading to a reduction of the surface-to-total ratio of a particular receptor or channel. For this reason, we focused our subsequent studies on Kir2.1. Overexpression of AGS3S, a short form of AGS3 lacking the N-terminal TPR domain (7), results in only a moderate increase in the surface density of Kir2.1 (Fig. 1F). Taken together, these data imply that AGS3 is involved in the trafficking of receptors and channels. Moreover, the TPR domain is required for AGS3-mediated enhancement of surface expression.
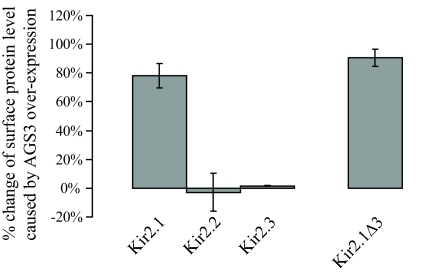
Effect of AGS3 on the Surface Level of Kir2.1 Is Independent of the Association Between Kir2.1 and PDZ Domain-Containing Proteins.
The close homolog of AGS3, mPins, has been shown to promote the surface expression of NMDA receptors via its interaction with PDZ domain-containing proteins (17). Because members of the Kir2 family can bind to PDZ proteins at their C termini (23), we wondered whether the effect of AGS3 on the trafficking of Kir2.1 is linked to PDZ proteins. To test this idea, we first compared the impacts of AGS3 and mPins on the surface level of Kir2.1 (Fig. 1F). Unlike AGS3, overexpressed mPins exerted little influence. Moreover, an elevated level of AGS3 failed to increase the surface expression of two other Kir2 members, Kir2.2 and Kir2.3 (Fig. 2), both of which have a PDZ-binding motif at their C termini. These results suggest that the presence of PDZ-binding motifs is not sufficient to render the trafficking of cargo sensitive to AGS3. We next determined whether the PDZ-binding motif is necessary for the effect of AGS3 by using a Kir2.1 mutant, Kir2.1Δ3, which lacks three residues of the C-terminal PDZ binding motif. Our data show that elevated AGS3 increases the surface level of the mutant channel to the same extent as it does the wild-type Kir2.1 (Fig. 2). In conclusion, our results demonstrate that AGS3 modulates the cargo trafficking in a way distinct from mPins.
Fig. 2.
Effects of AGS3 are independent of the PDZ-binding motifs of Kir2 channel proteins. Although AGS3 greatly increases the surface expression of Kir2.1, it does not significantly alter that of Kir2.2 or Kir2.3. Moreover, deletion of the PDZ-binding motif in Kir2.1Δ3 has no impact on the ability of AGS3 to stimulate the surface level of Kir2.1. Surface chemiluminescence measurements were conducted as described in Fig. 1. The values shown are the average of at least six independent experiments.
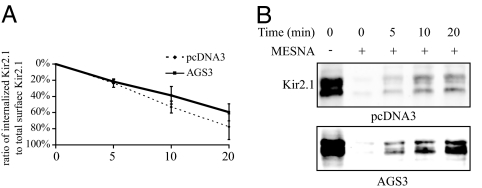
AGS3 Overexpression Results in a Modest Decrease in the Levels of Internalized Kir2.1 at Later Time Points.
The surface density of a receptor or channel can be controlled at various transport steps along the biosynthetic or endocytic pathways. To gain insight into the mechanism or mechanisms by which AGS3 regulates Kir2.1 trafficking, we investigated whether AGS3 affects the endocytic trafficking of Kir2.1. A biotin internalization assay (24) was used to compare the rates of internalization and/or recycling between control cells and cells overexpressing AGS3. The overexpression of AGS3 did not lead to an appreciable change in the relative amount of Kir2.1 internalized after 5 min (Fig. 3). However, at later time points (10 and 20 min), AGS3 overexpression led to a moderate decrease in the amount of channel remaining internalized when compared with the control sample. Thus, it seems likely that AGS3 overexpression alters the endocytic trafficking of Kir2.1.
Fig. 3.
Overexpression of AGS3 modestly affects the internalization or recycling of Kir2.1 at later, but not earlier, time points when compared with control. Surface proteins of COS7 cells overexpressing Kir2.1 and either AGS3 or pcDNA3 (control) were labeled by using the membrane-impermeant Sulfo-NHS-SS-Biotin (Pierce) and allowed to internalize for a period of 0, 5, 10, or 20 min. Remaining surface biotin was cleaved by using a membrane-impermeant reducing agent, mercaptoethanesulfonic acid (MESNA, 50 mM). Samples left untreated with MESNA at time 0 represent the total surface Kir2.1 protein. Proteins retaining their biotin tags after the MESNA treatment thus represented surface proteins that had become internalized but not yet recycled back to the plasma membrane. (A) Amounts of Kir2.1 remaining internalized after 0, 5, 10, and 20 min were normalized to total surface expression. Values shown are the average of four separate sets of Western blots. (B) A representative set of Western blots of the Kir2.1 protein internalization assay is shown.
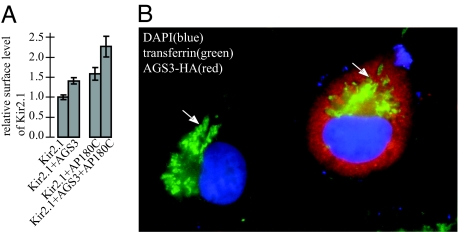
The Effect of AGS3 on the Surface Level of Kir2.1 Is Not Blocked After Inhibiting Clathrin-Mediated Internalization.
We next examined whether the effect of AGS3 on Kir2.1 is caused by its action at the clathrin-mediated internalization step. If AGS3 primarily regulates the clathrin-mediated endocytosis of Kir2.1 the increase in Kir2.1 surface levels observed in AGS3-overexpressing cells should be at least partially masked if the clathrin-mediated internalization of the channel protein is inhibited. To block the clathrin-mediated internalization of Kir2.1, we used a truncated mutant of AP180, AP180C (25). Compared with COS7 cells transfected with Kir2.1 plus a control vector, cells transfected with Kir2.1 plus AP180C displayed a 60% increase in channel proteins on the plasma membrane (Fig. 4A). This result indicates that a significant fraction of surface Kir2.1 is internalized via a clathrin-dependent mechanism. However, AGS3 increases the surface level of Kir2.1 to the same extent (≈40% increase) in the absence or presence of AP180C (Fig. 4A). Consistent with this notion, we found that clathrin-dependent internalization of transferrin receptors (26) proceeded normally in cells overexpressing AGS3 (Fig. 4B), suggesting that AGS3 does not facilitate the surface expression of Kir2.1 simply by inhibiting the clathrin-mediated internalization of Kir2.1.
Fig. 4.
Inhibition of clathrin-mediated internalization of Kir2.1 does not prevent the stimulation of channel surface expression by AG53 overexpression. (A) Surface expression of Kir2.1 was determined by chemiluminescence assay as in Fig. 1. Expression of AP180C efficiently inhibits the internalization of Kir2.1 (comparing Kir2.1 to Kir2.1+AP180C); however, it does not influence the AGS3-dependent increase of surface Kir2.1 (≈40% in either case, comparing Kir2.1+AGS3 with Kir2.1+AGS3+AP180C). Equal amounts of Kir2.1 were used for transfection in each experiment. Because a smaller amount of AGS3 was used for each transfection, the effects of AGS3 on the surface level of Kir2.1 appeared to be less compared with that in Figs. 1 or 2. (B) Exogenous biotinylated transferrin receptors are internalized normally and concentrated at the perinuclear recycling endosomes (indicated by arrows) in COS7 cells overexpressing AGS3.
Both Elevated and Decreased AGS3 Levels Alter Trans-Golgi Network (TGN) Structure and/or Protein Trafficking at the Level of the Trans-Golgi/TGN.
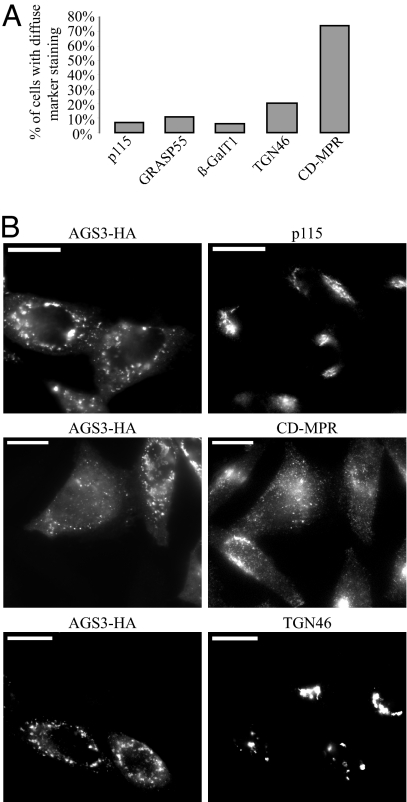
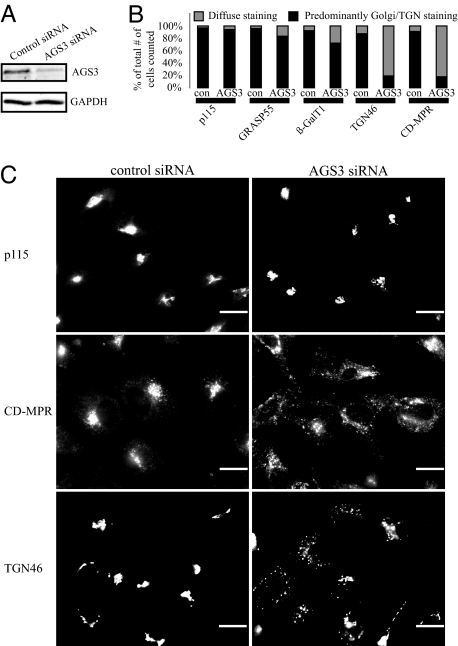
Because previous studies have revealed that a pool of AGS3 resides in the ER and Golgi compartments and one major interacting partner of AGS3, Gαi3, is concentrated at the Golgi apparatus (15, 16), we used immunoflouresence microscopy to investigate how AGS3 overexpression affected various components of the secretory pathway. An increased level of AGS3 did not cause any significant changes in the localizations of marker proteins for the ER (Calreticulin), the ER-to-Golgi COPII transport machinery (Sec13), the cis-Gogi network/cis-Golgi (p115), the medial-Golgi (GRASP55), the trans-Golgi/TGN (β-GalT1), and the lysosomes (Lamp1; Fig. 5 and unpublished observations), indicating that AGS3 overexpression does not disturb the general integrity of early secretory or lysosomal compartments. In contrast, the same treatment led to an extensive dispersal of TGN46 (Fig. 5), a cargo protein found at the trans-Golgi/TGN at steady state but that also shuttles among the TGN, the plasma membrane, and the endosomes (27), and CD-MPR, a protein trafficking primarily between the TGN and the endosomes (28). We then examined the localization of TGN46 and CD-MPR in HeLa cells where the endogenous AGS3 was depleted with siRNA. Compared with a nontargeting control siRNA, the commercial siRNA (Qiagen, Valencia, CA) targeting AGS3 mRNA (target sequence: CCGGGCGCTGGAATACCACAA) knocked down the AGS3 protein level by 80% (Western blot quantified by Li-Cor Odyssey), whereas GAPDH levels remained unchanged (Fig. 6A). Similar to overexpression, depletion of AGS3 had little or only modest influence on the markers of cis-Golgi network/cis-Golgi (p115), medial-Golgi (GRASP55), and β-GalT1, a trans-Golgi/TGN resident protein (Fig. 6B). Also like AGS3 overexpression, in AGS3 knockdown cells, TGN46 and CD-MPR were found to be greatly dispersed (Fig. 6C). This observation was specific to AGS3 depletion because cells treated with another siRNA targeting a different region of the AGS3 mRNA (target sequence: CCGCCGAGTACTACAAGAAGA) exerted a similar effect (data not shown). Our data imply that either the traffic in and/or out of the trans-Golgi/TGN or the integrity of the TGN are sensitive to the cellular AGS3 level.
Fig. 5.
Overexpression of AGS3 disrupts the localization of two endogenous TGN cargo proteins without affecting those of cis-Golgi/cis-Golgi network and medial-Golgi proteins. (A) COS7 cells overexpressing AGS3 were counted (n > 200, only cells highly overexpressing AG53 were counted) based on whether the staining of Golgi marker proteins had predominantly perinuclear or diffuse staining. (B) The localization of p115 is not significantly different in those cells highly overexpressing AGS3, whereas localization of the TGN cargo proteins TGN46 and CD-MPR are dramatically altered in cells with high levels of AGS3. Cells overexpressing and not overexpressing AGS3 were captured in the same field for comparison. (Scale bars: 20 μm.)
Fig. 6.
Similar to the overexpression of AGS3, depletion of AGS3 also leads to the specific dispersal of endogenous TGN cargo proteins. (A) The AGS3 siRNA used in this study efficiently knocked down the AGS3 protein level by 80% (Western blot quantified by the Li-Cor Odyssey Infrared Imaging System) without influencing that of GAPDH. (B) HeLa cells were counted (n > 200) based on whether the subcellular distributions of marker proteins were normal or diffuse. (C) p115 localization changes very little when cells are treated with the AGS3 siRNA, whereas the staining of the TGN cargo proteins, CD-MPR and TGN46, is much more diffuse when compared with control cells. (Scale bars: 20 μm.)
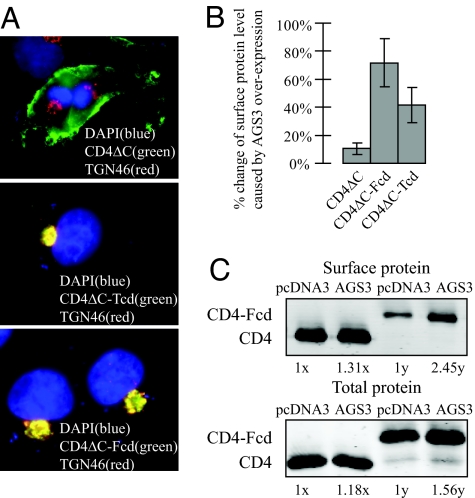
Elevated AGS3 Greatly Increases the Surface Expression of Two TGN-Enriched CD4 Reporters.
As a further test for our view that AGS3 primarily acts along the endocytic/TGN pathway or pathways, we assessed the effect of AGS3 overexpression on the surface levels of three CD4-derived reporter molecules, CD4ΔC, CD4ΔC-Fcd, and CD4ΔC-Tcd. CD4ΔC contains the extracellular region, transmembrane region, and 10 aa of the cytoplasmic region of CD4 and thus lacks any specific cytoplasmic trafficking motifs. It is stably localized to the plasma membrane when expressed in COS7 cells (Fig. 7A and ref. 29). CD4ΔC-Fcd and CD4ΔC-Tcd were made by fusing the cytoplasmic domain of Furin (Fcd) and TGN38 (Tcd) to CD4ΔC, respectively. Furin and TGN38 cycle between the TGN and plasma membrane, but both proteins enrich at the TGN at their steady state (30–32). This TGN enrichment occurs because, upon reaching the plasma membrane, Furin (31) and TGN38 (30, 32) are rapidly internalized and sent to the TGN from late and early endosomes, respectively. Previous studies have demonstrated that the C-terminal cytoplasmic domains of Furin and TGN38 are necessary and sufficient for their internalization and their subsequent endosome-to-TGN retrieval. Consistently both CD4ΔC-Fcd and CD4ΔC-Tcd mainly accumulated at the TGN (Fig. 7A and ref. 29). Because a much higher fraction of CD4ΔC-Fcd or CD4ΔC-Tcd cycles among the TGN, the plasma membrane, and the endosomes compared with CD4ΔC, we reasoned that if AGS3 functions at these compartments, its overexpression should impose a bigger effect on the surface density of CD4ΔC-Fcd or CD4ΔC-Tcd than that of CD4ΔC. Indeed, whereas overexpression of AGS3 did not greatly alter the surface level of CD4ΔC (<10% change), it greatly increased that of CD4ΔC-Fcd (by 60%) or CD4ΔC-Tcd (by 40%) as measured by a quantitative surface chemiluminescence assay (Fig. 7B). As an independent measurement we used surface biotinylation to measure the amount of surface expression of CD4ΔC and CD4ΔC-Fcd. AGS3 overexpression caused an increase in the total protein levels of CD4ΔC and CD4ΔC-Fcd (18% and 56%, respectively), and an increase in the surface protein levels of the two by 31% and 145%, respectively. These results indicate that an elevated level of AGS3 leads to an increase in the surface-to-total ratio of CD4ΔC-Fcd to a much higher degree than that of CD4ΔC and lend more support for a role for AGS3 in the TGN/plasma membrane/endosome trafficking pathway or pathways.
Fig. 7.
Elevated AGS3 greatly increases the surface expression of two TGN-enriched CD4-derived reporters. (A) Whereas CD4ΔC is efficiently expressed on the plasma membrane, both CD4ΔC-Tcd and CD4ΔC-Fcd primarily reside at the TGN. An anti-TGN46 antibody was used to label the TGN. (B) Surface chemiluminescence measurements of CD4ΔC, CD4ΔC-Fcd, and CD4ΔC-Tcd plasma surface expression were performed as described in Fig. 1. (C) A complementary surface biotinylation assay was used to determine surface levels of CD4ΔC and CD4ΔC-Fcd. 1x and 1y are arbitrary units and represent the total and surface levels of control samples (i.e., cells transfected with pcDNA3 only), respectively. AGS3 overexpression caused an increase in the CD4ΔC and CD4ΔC-Fcd total protein levels compared with controls (18% and 56%, respectively) and an increase in the surface protein levels of the two by 31% and 145%, respectively. Thus, the surface-to-total ratio of CD4ΔC-Fcd was stimulated by AGS3 to a much higher extent compared with that of CD4ΔC.
Discussion
Heterotrimeric G proteins have a well established role in relaying signals from GPCRs on the plasma membrane to downstream effectors in the cytosol. AGS3, originally identified in a screen for proteins that activate G protein signaling in a receptor-independent fashion, has been suggested to be involved in several different pathways, including drug-seeking behavior in animal models of addiction (11, 12). Given the well known role of receptor/channel trafficking in modulating neural plasticity and previously reported partial colocalization between AGS3 and markers of the ER and the Golgi apparatus (14), we were interested to see whether AGS3 could affect the plasma membrane expression of various GPCRs and channel proteins, and if so, whether this effect was a result of the regulation of protein trafficking.
By examining the impact of overexpressed AGS3 on the surface and total protein levels of a panel of plasma membrane receptors and channels, we found that AGS3 overexpression causes an increase in the total protein level of most, but not all, of the receptors and channels in transfected COS7 cells (Fig. 1B). This result is somewhat unexpected because cotransfection of two expression plasmids often leads to either little change or reduction of each expressed protein, presumably as a result of competition for the transcriptional or translational machinery. Despite the general stimulatory effect of AGS3 overexpression on the protein levels of receptors and channels, Kir2.1 is the only member whose surface expression is prominently enhanced (Fig. 1C). Because the surface level of Kir2.1 is increased to a significantly higher extent than that of the total Kir2.1 level (Fig. 1D), an elevated AGS3 expression appears to affect the trafficking of Kir2.1. Unlike its homolog mPins, the ability of AGS3 to influence Kir2.1 trafficking does not require the interaction of Kir2.1 with PDZ domain-containing proteins (Fig. 2). Instead, we have shown that internalization or recycling of Kir2.1 occurring over longer periods of time (10–20 min) is impacted by an increased level of AGS3 (Fig. 3), implying a role for AGS3 in the endocytic pathway. Moreover, overexpression and siRNA-mediated knockdown of AGS3 results in the specific dispersal of two endogenous TGN proteins cycling between the TGN and the plasma membrane or endosomes, TGN46 and CD-MPR, without altering the distributions of cis- and medial-Golgi markers (Figs. 5 and 6). This observation also points to a potential function of AGS3 in either the cargo trafficking into or out of the TGN or the structural integrity of the TGN. Additionally, CD4ΔC-Fcd and CD4ΔC-Tcd, two chimeric proteins that rapidly cycle between the TGN and the plasma membrane via endosomes (Fig. 7A), exhibit greatly increased plasma membrane levels in cells expressing an elevated amount of AGS3 (Fig. 7B). On the other hand, AGS3 overexpression exhibits little effect on the surface level of CD4ΔC, which stably resides on the plasma membrane. In sum, our studies have revealed a previously unrecognized role of AGS3 in regulating the trafficking of some plasma membrane proteins such as Kir2.1 between the cell surface and endosomes or TGN. It is important to point out that although our studies are focused mainly on Kir2.1, it remains possible that AGS3 also has a role in controlling the trafficking of other receptors and channels we examined (Fig. 1D). Another question to be addressed in the future regards the mechanism by which AGS3 affects the total protein level of receptors and channels.
Based on our studies, there are several modes through which AGS3 may influence membrane trafficking. One possibility is that AGS3 affects the TGN-to-plasma membrane transport of its cargo proteins, either via a biosynthetic pathway of newly synthesized proteins or a recycling pathway of internalized proteins through the TGN, as has been demonstrated for the recycling of several membrane proteins (30). The increase in the surface level of CD4ΔC-Tcd and CD4ΔC-Fcd (Fig. 7) and the dispersal of the TGN-associated cargo markers TGN46 and CD-MPR are consistent with such a function. In this model, the absence of a significant effect of AGS3 overexpression on the CD4ΔC surface level (Fig. 7) can be explained by a relative smaller pool of TGN-associated CD4ΔC at the steady state, compared with CD4ΔC-Tcd and CD4ΔC-Fcd. Moreover, the modest reduction seen during the later time points of Kir2.1 internalization can be caused by an increase of channel recycling through TGN in cells overexpressing AGS3. In this aspect, it would be interesting to see whether the impact of AGS3 persists when using Kir2.1 mutants with defective Golgi export (33). Alternatively, the preceding data can be explained if AGS3 were to specifically regulate a step in the endosome-to-plasma membrane recycling of surface proteins. Finally, we cannot rule out the possibility that AGS3 modulates the rate of endocytosis of its cargo. In this scenario, Kir2.1 might be internalized by two or more internalization pathways of differing kinetics (e.g., clathrin-dependent and clathrin-independent), with the AGS3-sensitive pathway operating mainly during the later time points under our experimental conditions. Such a model could explain why there was no difference seen in the level of internalized Kir2.1 between the AGS3 overexpression control samples in the early time point (Fig. 3) and why inhibition of clathrin-mediated internalization of Kir2.1 had little influence on the effect of AGS3 (Fig. 4). However, this model alone cannot readily account for the dispersal of CD-MPR, which is known to cycle primarily between the TGN and endosomes (28). Furthermore we were unable to detect the surface localization of CD-MPR after overexpressing or knocking down AGS3 (Figs. 5B and 6C). Future experiments will be required to distinguish among these possibilities.
Whereas our current study does not provide insight regarding the molecular mechanism by which AGS3 modulates membrane trafficking, it is noteworthy that heterotrimeric G proteins have been found to reside at many intracellular trafficking compartments such as the ER (34), the Golgi apparatus (15, 16), the secretory granules (35), the endosomes (36), and the cytoskeleton (37). Experiments using pharmacological tools and mutants of various Gα subunits suggest that G proteins are involved in the sorting of cargo proteins and the budding of transport carriers from the donor compartments (15, 16), as well as the fusion of transport carriers at the target compartments (35). Among them, the role of G proteins in regulating TGN export is best characterized (38). In this model, an unknown GPCR is assumed to activate a TGN-localized G protein, freeing the Gα and Gβγ (Gβ1γ2 and Gβ3γ2) subunits. The Gβγ subunit promotes a signaling cascade, leading to the recruitment of multiple effector proteins to stimulate the fission of cargo-containing carriers bound for the plasma membrane. Based on our findings, one intriguing possibility would be that AGS3 can modulate the protein transport at the Golgi apparatus by sequestering the Gα subunit, inhibiting the reassociation of Gβ1γ2 and Gβ3γ2 with Gα, thus enhancing the Gβγ-mediated Golgi export. Our observations showing increased surface expression of Kir2.1, CD4ΔC-Tcd, and CD4ΔC-Fcd upon AGS3 overexpression are consistent with this possibility. In this scenario, the dispersal of TGN46 and CD-MPR could be the consequence of an imbalance in the TGN traffic.
Whereas AGS3 and its close homolog mPins are both abundantly expressed in the brain, they display different subcellular distributions and responses to stimuli, suggesting distinct functions (14). Several previous studies have implicated mPins in tuning synaptic plasticity via regulating the trafficking of NMDA receptor or the activity of G protein-activated inwardly rectifying potassium channels in hippocampal neurons (17, 39). Our current findings raise the exciting possibility that AGS3 and mPins may constitute a family of important modulators of neural plasticity.
Materials and Methods
DNA Constructs and Reagents.
CD4ΔC, CD4ΔC-Fcd, and CD4ΔC-Tcd constructs have been described (29). AGS3 and all receptors and channels were cloned into the expression vector pcDNA3 (Invitrogen). The antibodies used in this study were: monoclonal anti-HA (HA.11; Covance, Richmond, CA), CD4 (Chemicon), p115 (BD Transduction), GRASP55 (a gift from F. Barr, Cancer Research Center, University of Liverpool, Liverpool, U.K.), β-GalT1 (a gift from U. Mandel, University of Copenhagen, Copenhagen), TGN46 (a gift from S. Ponnambalam, University of Dundee, Dundee, U.K.), and CD-MPR (Hybridoma Bank). A polyclonal antibody raised previously against the N-terminal portion of Kir2.1 (peptide sequence: A V A N G F G N G K S K V H T R Q Q K) was also used.
Cell Culture and Transfection.
COS7 or HeLa cells were cultured in Advanced DMEM (GIBCO) supplemented with 4% FBS, 2 mM glutamine, and 1× penicillin-streptomycin (Cellgro). FuGENE HD (Roche, Indianapolis, IN) or Lipofectamine 2000 (Invitrogen) was used for transfecting cells with DNA constructs or siRNA, respectively.
Quantitative Surface Chemiluminescence Assay.
We followed a surface chemiluminescence protocol as described (19, 29, 40).
SDS/PAGE and Western Blot Analysis.
Protein samples were separated by SDS/PAGE, and Western blot was performed as described (29). Quantitation was performed on an infrared imaging system (Odyssey).
Biotinylated Transferrin Internalization Assay.
Biotinylated transferring (Jackson ImmunoResearch) internalization was performed as described (29).
Surface Biotinylation Assay.
Surface biotinylation was performed according to the manufacturer's instructions with the Cell Surface Protein Isolation Kit (Pierce, Rockford, IL) with Sulfo-NHS-SS-Biotin.
Biotin Internalization Assay.
The biotin internalization assay was performed as described (24).
Immunostaining Assay.
Immunostaining of HeLa and COS7 cells was performed as described (29).
Acknowledgments
We thank the individuals who provided antibodies and/or reagents (see Materials and Methods).
Abbreviations
- AG53
activator of G protein signaling 3
- ER
endoplasmic reticulum
- GPCR
G protein-coupled receptor
- TGN
trans-Golgi network.
Footnotes
The authors declare no conflict of interest.
References
- 1.Takesono A, Cismowski MJ, Ribas C, Bernard M, Chung P, Hazard S, 3rd, Duzic E, Lanier SM. J Biol Chem. 1999;274:33202–33205. doi: 10.1074/jbc.274.47.33202. [DOI] [PubMed] [Google Scholar]
- 2.Blatch GL, Lassle M. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 3.Willard FS, Kimple RJ, Siderovski DP. Annu Rev Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- 4.Peterson YK, Bernard ML, Ma H, Hazard S, 3rd, Graber SG, Lanier SM. J Biol Chem. 2000;275:33193–33196. doi: 10.1074/jbc.C000509200. [DOI] [PubMed] [Google Scholar]
- 5.De Vries L, Fischer T, Tronchere H, Brothers GM, Strockbine B, Siderovski DP, Farquhar MG. Proc Natl Acad Sci USA. 2000;97:14364–14369. doi: 10.1073/pnas.97.26.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Natochin M, Lester B, Peterson YK, Bernard ML, Lanier SM, Artemyev NO. J Biol Chem. 2000;275:40981–40985. doi: 10.1074/jbc.M006478200. [DOI] [PubMed] [Google Scholar]
- 7.Pizzinat N, Takesono A, Lanier SM. J Biol Chem. 2001;276:16601–16610. doi: 10.1074/jbc.M007573200. [DOI] [PubMed] [Google Scholar]
- 8.Pattingre S, De Vries L, Bauvy C, Chantret I, Cluzeaud F, Ogier-Denis E, Vandewalle A, Codogno P. J Biol Chem. 2003;278:20995–21002. doi: 10.1074/jbc.M300917200. [DOI] [PubMed] [Google Scholar]
- 9.Pattingre S, Petiot A, Codogno P. Methods Enzymol. 2004;390:17–31. doi: 10.1016/S0076-6879(04)90002-X. [DOI] [PubMed] [Google Scholar]
- 10.Sanada K, Tsai LH. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Bowers MS, McFarland K, Lake RW, Peterson YK, Lapish CC, Gregory ML, Lanier SM, Kalivas PW. Neuron. 2004;42:269–281. doi: 10.1016/s0896-6273(04)00159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao L, McFarland K, Fan P, Jiang Z, Inoue Y, Diamond I. Proc Natl Acad Sci USA. 2005;102:8746–8751. doi: 10.1073/pnas.0503419102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernard ML, Peterson YK, Chung P, Jourdan J, Lanier SM. J Biol Chem. 2001;276:1585–1593. doi: 10.1074/jbc.M005291200. [DOI] [PubMed] [Google Scholar]
- 14.Blumer JB, Chandler LJ, Lanier SM. J Biol Chem. 2002;277:15897–15903. doi: 10.1074/jbc.M112185200. [DOI] [PubMed] [Google Scholar]
- 15.Stow JL, de Almeida JB, Narula N, Holtzman EJ, Ercolani L, Ausiello DA. J Cell Biol. 1991;114:1113–1124. doi: 10.1083/jcb.114.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barr FA, Leyte A, Mollner S, Pfeuffer T, Tooze SA, Huttner WB. FEBS Lett. 1991;294:239–243. doi: 10.1016/0014-5793(91)81438-e. [DOI] [PubMed] [Google Scholar]
- 17.Sans N, Wang PY, Du Q, Petralia RS, Wang YX, Nakka S, Blumer JB, Macara IG, Wenthold RJ. Nat Cell Biol. 2005;7:1179–1190. doi: 10.1038/ncb1325. [DOI] [PubMed] [Google Scholar]
- 18.Margeta-Mitrovic M, Jan YN, Jan LY. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 19.Zerangue N, Schwappach B, Jan YN, Jan LY. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 20.Ma D, Zerangue N, Lin YF, Collins A, Yu M, Jan YN, Jan LY. Science. 2001;291:316–319. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- 21.Ma D, Zerangue N, Raab-Graham K, Fried SR, Jan YN, Jan LY. Neuron. 2002;33:715–729. doi: 10.1016/s0896-6273(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 22.Tsao PI, von Zastrow M. J Biol Chem. 2000;275:11130–11140. doi: 10.1074/jbc.275.15.11130. [DOI] [PubMed] [Google Scholar]
- 23.Nehring RB, Wischmeyer E, Doring F, Veh RW, Sheng M, Karschin A. J Neurosci. 2000;20:156–162. doi: 10.1523/JNEUROSCI.20-01-00156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morimoto S, Nishimura N, Terai T, Manabe S, Yamamoto Y, Shinahara W, Miyake H, Tashiro S, Shimada M, Sasaki T. J Biol Chem. 2005;280:2220–2228. doi: 10.1074/jbc.M406906200. [DOI] [PubMed] [Google Scholar]
- 25.Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 26.Klausner RD, Van Renswoude J, Ashwell G, Kempf C, Schechter AN, Dean A, Bridges KR. J Biol Chem. 1983;258:4715–4724. [PubMed] [Google Scholar]
- 27.Ponnambalam S, Girotti M, Yaspo ML, Owen CE, Perry AC, Suganuma T, Nilsson T, Fried M, Banting G, Warren G. J Cell Sci. 1996;109:675–685. doi: 10.1242/jcs.109.3.675. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh P, Dahms NM, Kornfeld S. Nat Rev Mol Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 29.Gong Q, Weide M, Huntsman C, Xu Z, Jan LY, Ma D. J Biol Chem. 2007;282:13087–13097. doi: 10.1074/jbc.M700767200. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh RN, Mallet WG, Soe TT, McGraw TE, Maxfield FR. J Cell Biol. 1998;142:923–936. doi: 10.1083/jcb.142.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, Thomas G. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 32.Mallet WG, Maxfield FR. J Cell Biol. 1999;146:345–359. doi: 10.1083/jcb.146.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofherr A, Fakler B, Klocker N. J Cell Sci. 2005;118:1935–1943. doi: 10.1242/jcs.02322. [DOI] [PubMed] [Google Scholar]
- 34.Schwaninger R, Plutner H, Bokoch GM, Balch WE. J Cell Biol. 1992;119:1077–1096. doi: 10.1083/jcb.119.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang J, Nishimoto I, Okamoto T, Regazzi R, Kiraly C, Weller U, Wollheim CB. EMBO J. 1995;14:3635–3644. doi: 10.1002/j.1460-2075.1995.tb00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Dyke RW. BMC Physiol. 2004 doi: 10.1186/1472-6793-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis JM, Woolkalis MJ, Gerton GL, Smith RM, Jarett L, Manning DR. Cell Regul. 1991;2:1097–1113. doi: 10.1091/mbc.2.12.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diaz Anel AM, Malhotra V. J Cell Biol. 2005;169:83–91. doi: 10.1083/jcb.200412089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiser O, Qian X, Ehlers M, Ja WW, Roberts RW, Reuveny E, Jan YN, Jan LY. Neuron. 2006;50:561–573. doi: 10.1016/j.neuron.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 40.Margeta-Mitrovic M. Methods. 2002;27:311–317. doi: 10.1016/s1046-2023(02)00088-9. [DOI] [PubMed] [Google Scholar]