Abstract
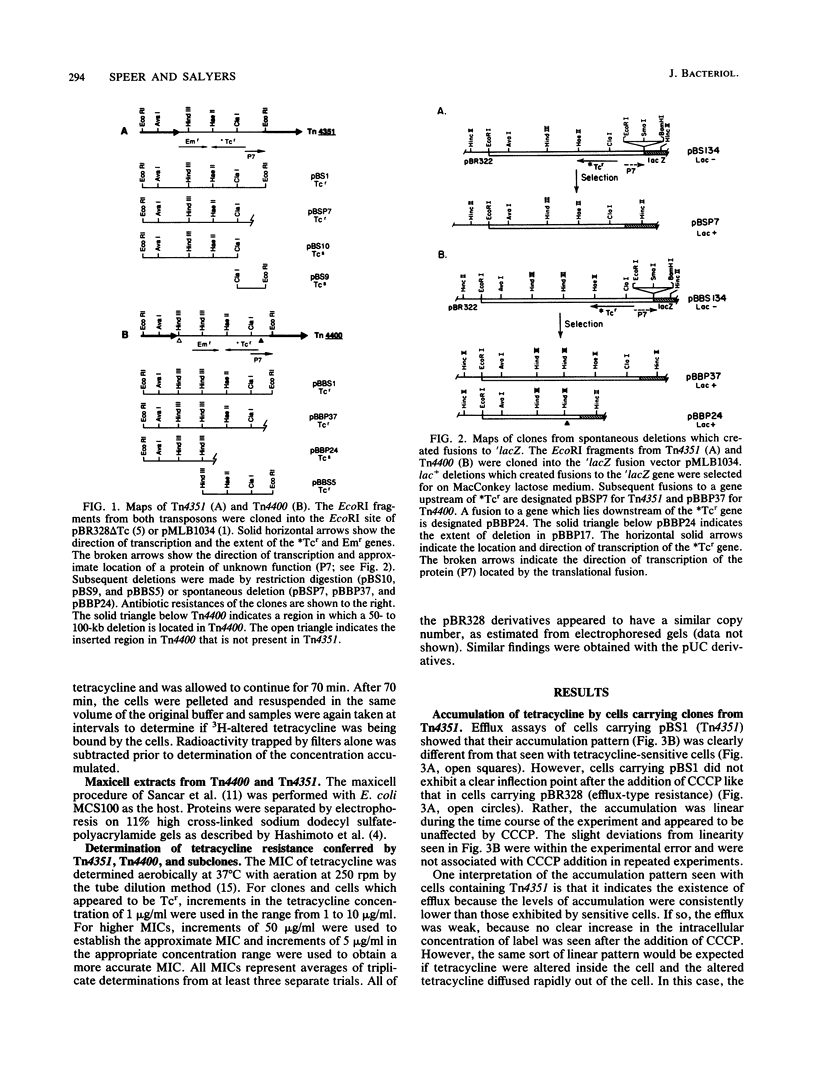
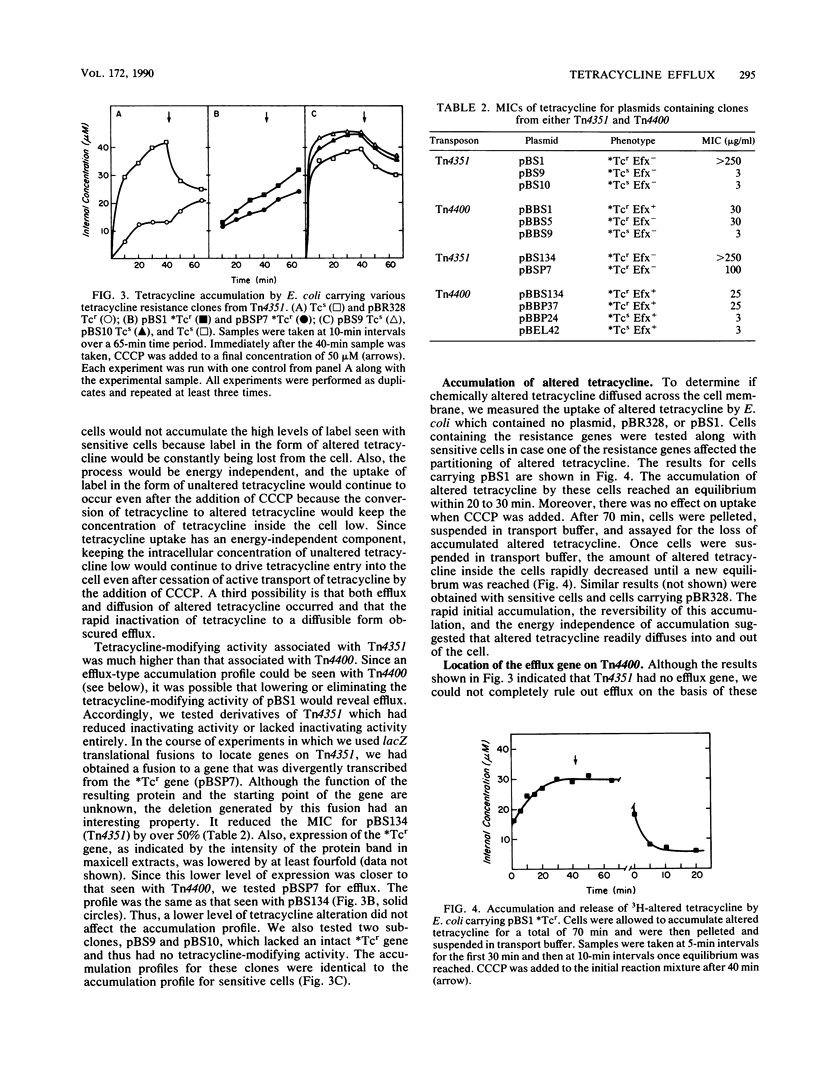
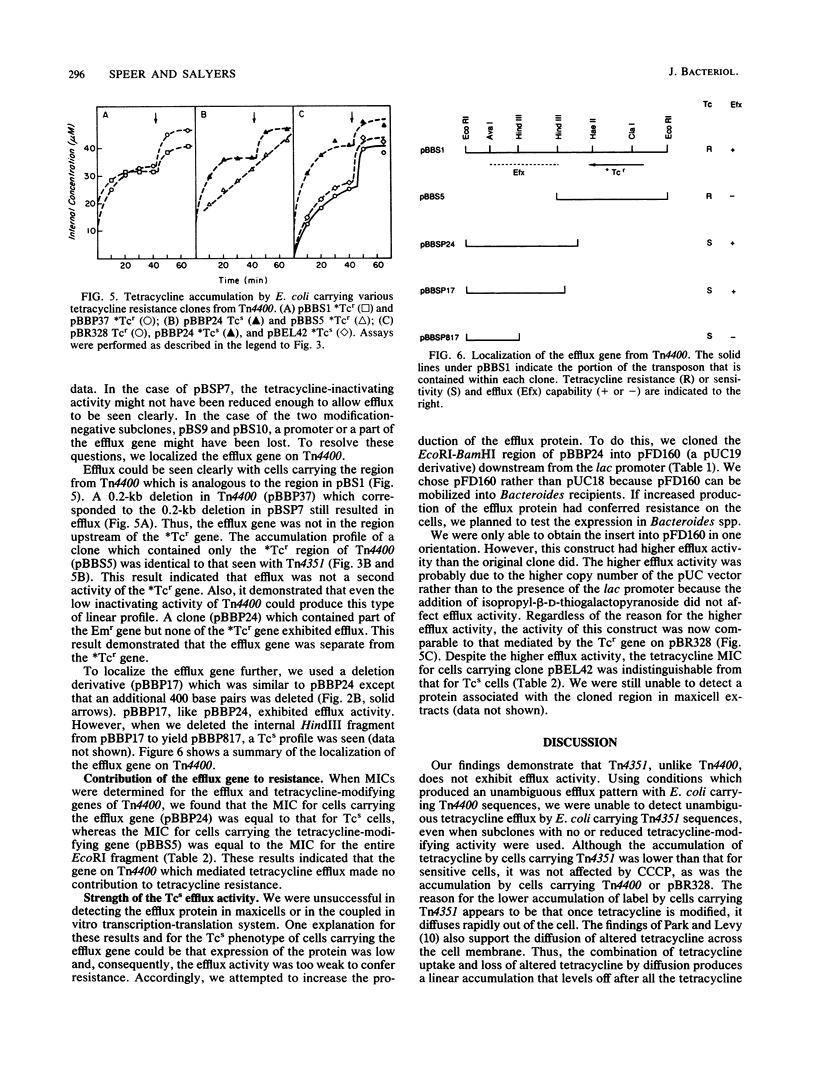
Previously, we demonstrated that the Bacteroides transposon Tn4351, which confers tetracycline resistance only on aerobically grown Escherichia coli, carries a gene that codes for a tetracycline-inactivating enzyme (B. S. Speer and A. A. Salyers, J. Bacteriol. 170:1423-1429, 1988). However, Park et al. (B. H. Park, M. Hendricks, M. H. Malamy, F. P. Tally, and S. B. Levy, Antimicrob. Agents Chemother. 31:1739-1743, 1987) showed that E. coli carrying a closely related transposon, Tn4400, exhibits energy-dependent efflux of tetracycline as well as tetracycline-inactivating activity (B. H. Park and S. B. Levy, Antimicrob. Agents Chemother. 32:1797-1800, 1988). This result raised the question of whether efflux or inactivation or a combination of the two was necessary for resistance conferred by both transposons. We showed that cells carrying Tn4351 did not exhibit the clear-cut efflux activity seen with cells carrying Tn4400 but rather exhibited a tetracycline accumulation profile which could be explained solely on the basis of inactivation of tetracycline in the cytoplasm and rapid diffusion of altered tetracycline out of the cell. Additionally, we were able to clone the efflux and tetracycline-modifying genes of Tn4400 separately. The region carrying the efflux gene spanned one of the two regions in which Tn4400 differs from Tn4351. A clone containing the corresponding region of Tn4351 did not exhibit efflux. Thus, it appears that Tn4351 does not have the efflux gene and that efflux makes no contribution to the resistance conferred by Tn4351. The MIC for cells carrying the subclone from Tn4400 that contained only the gene for tetracycline inactivation was the same that for cells carrying both the inactivation and efflux genes. Cells carrying only the gene for tetracycline efflux were tetracycline sensitive. This was true even when the efflux gene was on a high-copy-number plasmid which increased the level of efflux to that associated with the Tcr gene on pBR328. These results indicate that efflux activity does not contribute significantly to the tetracycline resistance conferred by Tn4400.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman M. L., Jackson D. E. Selection of lac gene fusions in vivo: ompR-lacZ fusions that define a functional domain of the ompR gene product. J Bacteriol. 1984 Aug;159(2):750–756. doi: 10.1128/jb.159.2.750-756.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayolle F., Privitera G., Sebald M. Tetracycline transport in Bacteroides fragilis. Antimicrob Agents Chemother. 1980 Oct;18(4):502–505. doi: 10.1128/aac.18.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Expression in Escherichia coli of cryptic tetracycline resistance genes from bacteroides R plasmids. Plasmid. 1984 May;11(3):248–252. doi: 10.1016/0147-619x(84)90031-3. [DOI] [PubMed] [Google Scholar]
- Hashimoto F., Horigome T., Kanbayashi M., Yoshida K., Sugano H. An improved method for separation of low-molecular-weight polypeptides by electrophoresis in sodium dodecyl sulfate-polyacrylamide gel. Anal Biochem. 1983 Feb 15;129(1):192–199. doi: 10.1016/0003-2697(83)90068-4. [DOI] [PubMed] [Google Scholar]
- Matthews B. G., Guiney D. G. Characterization and mapping of regions encoding clindamycin resistance, tetracycline resistance, and a replication function on the Bacteroides R plasmid pCP1. J Bacteriol. 1986 Aug;167(2):517–521. doi: 10.1128/jb.167.2.517-521.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L., Petrucci R. E., Jr, Levy S. B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. H., Hendricks M., Malamy M. H., Tally F. P., Levy S. B. Cryptic tetracycline resistance determinant (class F) from Bacteroides fragilis mediates resistance in Escherichia coli by actively reducing tetracycline accumulation. Antimicrob Agents Chemother. 1987 Nov;31(11):1739–1743. doi: 10.1128/aac.31.11.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. H., Levy S. B. The cryptic tetracycline resistance determinant on Tn4400 mediates tetracycline degradation as well as tetracycline efflux. Antimicrob Agents Chemother. 1988 Dec;32(12):1797–1800. doi: 10.1128/aac.32.12.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Barber R. D., Salyers A. A. Cloning and characterization of a Bacteroides conjugal tetracycline-erythromycin resistance element by using a shuttle cosmid vector. J Bacteriol. 1989 Mar;171(3):1294–1302. doi: 10.1128/jb.171.3.1294-1302.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J. Development and use of cloning systems for Bacteroides fragilis: cloning of a plasmid-encoded clindamycin resistance determinant. J Bacteriol. 1985 Oct;164(1):294–301. doi: 10.1128/jb.164.1.294-301.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Gonda M. A. Comparison of the transposon-like structures encoding clindamycin resistance in Bacteroides R-plasmids. Plasmid. 1985 May;13(3):182–192. doi: 10.1016/0147-619x(85)90041-1. [DOI] [PubMed] [Google Scholar]
- Speer B. S., Salyers A. A. Characterization of a novel tetracycline resistance that functions only in aerobically grown Escherichia coli. J Bacteriol. 1988 Apr;170(4):1423–1429. doi: 10.1128/jb.170.4.1423-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer B. S., Salyers A. A. Novel aerobic tetracycline resistance gene that chemically modifies tetracycline. J Bacteriol. 1989 Jan;171(1):148–153. doi: 10.1128/jb.171.1.148-153.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]


