Abstract
To respond to potential adverse exposures properly, health care providers need accurate indicators of exposure levels. The indicators are particularly important in the case of acetaminophen (APAP) intoxication, the leading cause of liver failure in the U.S. We hypothesized that gene expression patterns derived from blood cells would provide useful indicators of acute exposure levels. To test this hypothesis, we used a blood gene expression data set from rats exposed to APAP to train classifiers in two prediction algorithms and to extract patterns for prediction using a profiling algorithm. Prediction accuracy was tested on a blinded, independent rat blood test data set and ranged from 88.9% to 95.8%. Genomic markers outperformed predictions based on traditional clinical parameters. The expression profiles of the predictor genes from the patterns extracted from the blood exhibited remarkable (97% accuracy) transtissue APAP exposure prediction when liver gene expression data were used as a test set. Analysis of human samples revealed separation of APAP-intoxicated patients from control individuals based on blood expression levels of human orthologs of the rat discriminatory genes. The major biological signal in the discriminating genes was activation of an inflammatory response after exposure to toxic doses of APAP. These results support the hypothesis that gene expression data from peripheral blood cells can provide valuable information about exposure levels, well before liver damage is detected by classical parameters. It also supports the potential use of genomic markers in the blood as surrogates for clinical markers of potential acute liver damage.
Keywords: acetaminophen, hepatotoxicity, microarray, prediction, genomics
Liver injury is the most commonly observed adverse effect in response to many environmental exposures. The incidence of xenobiotic-induced hepatic injury is estimated to be ≈14/100,000 inhabitants of Western countries (1). Among these individuals, acetaminophen (APAP) is responsible for the majority of clinical cases that present with acute liver failure (1). Recently, it has been shown that treatment with recommended doses of APAP frequently produces liver injury in healthy adults (2). There is an effective antidote for APAP intoxication, N-acetyl cysteine, that can minimize liver injury (3). However, 50% of overdoses are unintentional, and patients may present with undetectable levels of APAP. The early detection of APAP liver injury and determination of prognosis at presentation are critical to clinicians but can be challenging. Serum markers are not very sensitive and are poor predictors of outcome (4). Liver biopsies to obtain material for histopathological evaluations are invasive and are connected with a significant risk for the patient (5, 6). Serum APAP levels can be low or undetectable once liver injury occurs (2). Thus, there is a need for novel diagnostic and prognostic indicators using biomaterial that can be obtained with minimal invasion.
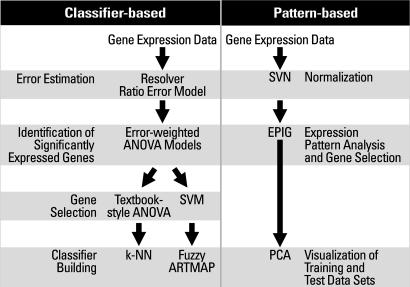
Gene expression technology using microarrays allows analysis of thousands of genes in parallel. A challenge in these studies is to identify signature patterns of genes that allow prediction of classes of samples with a high degree of accuracy. In this study, we tested the hypothesis that it is possible to predict acute exposure to harmful levels of an agent based solely on gene expression data obtained from blood cells. Prediction algorithms used classifiers and a pattern-based method (Fig. 1). Emphasis was given to the utilization of genomic markers for times after exposure that preceded clinical signs of injury. We used a rat model to generate a training data set consisting of genomic, clinical chemistry, histopathology, and hematology data. These measurements were analyzed for criteria that allowed discrimination of exposure levels that would not be injurious to the liver, i.e., “nontoxic” or “subtoxic,” from levels that would be expected to result in serious liver injury, i.e., “toxic.” Subsequently, those criteria were used to predict the exposure level of independent, blinded test samples. The accuracy of prediction was compared between the various measurements.
Fig. 1.
Workflow to predict the exposure level of the samples. The steps in the classifier-based and pattern-based approaches are shown.
Our study demonstrates that the accuracy of prediction with gene expression data is significantly better than prediction based on clinical chemistry, hematology, or histopathology. Our results also demonstrate that blood gene expression data are sufficient to predict exposure to harmful levels of APAP. In addition, analysis of human samples revealed separation of APAP-intoxicated patients from control individuals based on blood expression levels of human orthologs of the rat discriminatory genes. The results suggest that such a gene expression signature could be useful and supports further testing to determine the extent that such indicators can be translated into the clinical setting for surveying individuals presenting with APAP intoxication.
Results
We developed training and test data sets to test the hypothesis that genomic analysis of whole blood RNA could allow prediction of levels of APAP exposure, and to optimize and test the performance of prediction algorithms. The training set included male Fisher rats treated with 0, 150, 1,500, or 2,500 mg/kg APAP by oral gavage, killed 6, 12, or 24 h after exposure. The test data consisted of rats treated with 0, 150, 1,500, or 2,000 mg/kg APAP by oral gavage, killed 3, 6 or 24 h after treatment. Both sets included gene expression data from animals exposed to subtoxic (150 mg/kg) and toxic (1,500 and 2,000 or 2,500 mg/kg) doses of APAP hybridized against a time-matched vehicle control. Additionally, the test data set contained genomic data from animals that had received treatment with vehicle-only (nontoxic) hybridized against the vehicle-only controls. The test data set was evaluated in a blinded fashion.
Clinical Chemistry Parameters Lack Discriminating Sensitivity.
Based on alanine aminotransferase (ALT) and sorbitol dehydrogenase (SDH) data obtained from the training set, we set threshold values that would indicate exposure to a toxic versus “subtoxic/nontoxic” dose of APAP. Because clinical chemistry measurements after 6 and 12 h were indistinguishable between the dose groups, we could use only the 24 h values for this exercise. We determined the range of activity levels for ALT and SDH in the training set [Table 1 and supporting information (SI) Fig. 3]. Based on those ranges of activity levels, we assigned the animals of the test group using the following criteria: (i) if at least one of the two clinical chemistry measurements fell within the range of activity for either subtoxic/nontoxic or toxic dose, the animal was predicted as belonging to the respective group; (ii) if one measurement fell within the subtoxic/nontoxic dose range and the other into the toxic dose range, the animal was predicted toxic; (iii) if neither parameter fell into a predefined range of activity, the animal was classified as not predictable. By using those criteria on all animals in the test set, only 45 of 72 animals were predicted correctly, resulting in an accuracy of prediction of 62.5% [Table 2 and SI Table 4].
Table 1.
Shown are the lowest and highest values of the indicated clinical chemistry parameters in either the 0 and 150 mg/kg APAP groups at 24 h (sub/nontoxic dose) or the 1,500 and 2,500 mg/kg APAP groups at 24 h (toxic dose)
| Dose | Activity, units/liter |
|
|---|---|---|
| ALT | SDH | |
| Subtoxic/nontoxic | 41–59 (48.5) | 1.8–9.2 (6.2) |
| Toxic | 1,160–15,400 (10,420) | 10.3–1,400 (720) |
We defined the range these values span as predictive areas for toxicity of the given parameter. The numbers in parenthesis indicate the median.
Table 2.
Summary of prediction accuracies of the test data set with different predictive methods trained on the training set
| Clinical chemistry | Hematology | Histopathology |
Microarray bioinformatics analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Pathologist 1 | Pathologist 2 | Pathologist 3 | EPIG | k-NN | DME ANOVA | DCE ANOVA | |||
| Accuracy of prediction, % | 62.5 | 77.8 | 75 | 66.7 | 70.8 | 91.6 | 95.8 | 95.8 | 88.9 |
| (75) | (71) | (86) | (82) | (86) | (79.2) | (91.2) | (95.8) | (91.2) | |
| [97.2] | [54.2] | [58.3] | [56.9] | ||||||
In parentheses are displayed the prediction accuracies for the 24 h time point only of the test data set; in brackets are displayed the prediction accuracies for the liver gene expression data of the test set based on blood classifiers.
Histopathological Evaluation Shows Limited Accuracy of Prediction at Early Time Points After Treatment.
Board-certified veterinary pathologists (n = 3) extensively evaluated liver histologic slides from the training set to gauge the range, degree, and extent of hepatocyte necrosis and degeneration after toxic, subtoxic, and nontoxic APAP exposure. The same pathologists each evaluated the liver of the animals in the test set in a blinded fashion. To test the predictive power of histopathological evaluation, they were to determine, based solely on histologic evidence, which animals had received a subtoxic/nontoxic dose or a toxic dose of APAP. The overall accuracy of prediction varied from 66.7% to 75% between the pathologists (Table 2). The majority of missed calls were due to false negative results at early time points (3 and 6 h) after toxic exposure (SI Table 5).
Blood Cell Counts Allow Limited Prediction of Toxic Treatment.
Based on lymphocyte and neutrophil counts retrieved from the training set, threshold values were set that would allow prediction of unknown samples from the blood test set as either subtoxic/nontoxic or toxic. Because of the strong diurnal variation of the baseline counts (SI Table 6), it was necessary to determine a neutrophil to lymphocyte ratio for each animal and establish thresholds for those ratios (Table 3). Thus, animals in this set were grouped into either subtoxic/nontoxic or toxic depending on their neutrophil-to-lymphocyte ratio or not-determinable if they fell outside those thresholds. By using these criteria, 56 of 72 animals were predicted correctly: an accuracy of 77.8% (Table 2).
Table 3.
The lowest and highest values of the neutrophil/lymphocyte cell count ratio in either the 0 and 150 mg/kg APAP group at 6, 12, or 24 h (subtoxic/nontoxic) or the 1,500 and 2,500 mg/kg APAP group at 6, 12, or 24 h (toxic)
| Dose | Neutrophil/lymphocyte cell count ratio |
|---|---|
| Subtoxic/nontoxic | 0.172–0.626 |
| Toxic | 0.75–3.42 |
The range these values span we defined as predictive areas for toxicity of the given parameter.
Selection of the Predictor Genes.
To identify the genes with expression values in the blood training samples that vary significantly between subtoxic- and toxic-dosed samples, two ANOVA models were constructed. In one case, a main effect for the dose exposure was modeled, and, in the other case, dose and time main effects were confounded. By using the dose main effect (DME) ANOVA with Bonferroni correction (P value = 0.05) for multiple comparisons, 152 genes were identified as significantly different between comparisons of the dose administered to the samples (SI Table 7). When dose and time were confounded, the dose confounded effect (DCE) ANOVA without a correction for multiple comparisons yielded 264 genes that were significantly different between contrasts of subtoxic-dosed and toxic-dosed samples (SI Table 7). Applying a Bonferroni correction for multiple comparisons with the DCE ANOVA model analysis yielded few (n = 9) significant genes.
The gene expression ratio values for the 152 genes and from the training samples were analyzed by a k-nearest neighbors (k-NN) classifier with 10-fold cross validation and a multicategory support vector machine (MC-SVM) using leave-one-out cross validation (LOOCV) to select genes which had a high accuracy of prediction. A >95% accuracy of prediction was obtained by using the top 35 significant genes from the DME ANOVA model and k-NN classifier (SI Table 7). A 97.1% accuracy prediction rate was achieved by using 20 genes selected as optimal for prediction via the MC-SVM. Similarly, a 97.1% accuracy prediction rate was achieved by using 20 genes selected as optimal for prediction via the MC-SVM on the 264 genes from the DCE ANOVA (SI Table 7). In the MC-SVM cases, a single rat (no. 3338), analyzed 12 h after being dosed with 2,500 mg/kg APAP, was predicted incorrectly as a subtoxic-dosed sample. Principal component analysis (PCA) of the training samples using the 152 genes selected by the DME ANOVA and plotting the first three principal components shows that rat no. 3338 situated in dimensional space very close to the subtoxic dose samples (SI Fig. 4a).
Building Classifiers for Prediction of the Blinded Samples.
By using the 35 predictor genes from the DME ANOVA model and k-NN classifier, a 95.8% accuracy of prediction of the blood test samples was achieved (Table 2). All of the toxic dose samples were predicted correctly independent of time (SI Table 8). However, the predicted subtoxic/nontoxic group contained three toxic-dosed samples (two 1,500 mg/kg and one 2,000 mg/kg). By using both sets of 20 predictor genes selected by the MC-SVM, two classifiers were constructed on the training data using a fuzzy adaptive resonance theory map (Fuzzy ARTMAP) neural network (7, 8). LOOCV of the training data at 0.01 increments over the range of the vigilance parameter (from 0 to 1), indicated that a vigilance parameter value of 0.2 was sufficient for maximal accuracy (data not shown). To predict the class of dose administered to the rats, the sets of 20 genes selected as predictors were used to compile the gene expression ratio values from the test set samples. The gene expression data were analyzed by using the Fuzzy ARTMAP neural network with the vigilance parameter set at 0.2, and the gene expression ratio values from the training data were used to construct the classifier. The accuracy of the prediction results are shown in Table 2. Compared with the true dose classification, the 20 predictor genes from the DME ANOVA had a higher accuracy of prediction (95.8%) than the 20 predictor genes from the DCE ANOVA (88.9%) (SI Table 8).
Extracting Gene Expression Patterns in the Blood Data for Prediction of Exposure.
By using the extracting patterns and identifying genes (EPIG) approach (9), eight distinct patterns of gene expression were obtained from the training data (SI Fig. 4b). Pattern 1 has a clear separation of the subtoxic/nontoxic and the toxic-dosed samples based on the expression of the genes above or below a log base 2 ratio value of ≈−0.3. From the eight patterns, 248 genes were selected by EPIG and included in the signature list for prediction (SI Table 7).
PCA of the training and the test samples simultaneously using the 248 genes in the signature list was performed to predict the classes of the samples. When test samples were projected into 3D PCA space, the visualized closeness to either class of the training samples was judged, and membership to a class was predicted (SI Fig. 5a), resulting in a 91.6% accuracy (Table 2 and SI Table 8).
Inflammatory Processes as the Most Significant Biological Discriminator Between Subtoxic/Nontoxic and Toxic Exposures.
The gene selection methods used in this work yielded from 20 (DME and DCE ANOVAs with SVM), 35 (ANOVA/k-NN), and 248 (EPIG) predictor genes. Ten genes were common to the four methods (SI Table 7). The union of all predictor genes resulted in a total of 270 genes. The main Gene Ontology (GO) categories impacted by these genes involve the activation of immune or inflammatory responses against an external stimulus. Examples of overrepresented categories are defense response, immune response, response to stress, regulation of phagocytosis, regulation of endocytosis, response to bacteria, and inflammatory response. Analyzing the GO categories impacted by the individual predictor gene sets resulted in the following. EPIG genes indicated similar immune response activation as with the analysis of the gene union. The highest scores for the k-NN genes were obtained for specific immune response GO categories (MHC protein complex, immunological synapse), in agreement with the major theme of immune or inflammatory responses. The most significant GO categories affected in both DCE ANOVA and DME ANOVA gene sets reflect an interleukin-1-mediated inflammatory response, as did the 10 genes shared by all of the predictive gene lists.
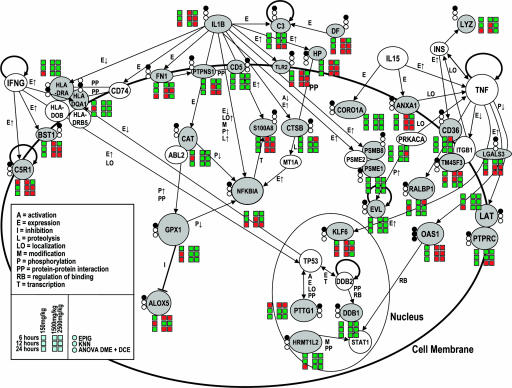
GO analysis of all lists identified genes involved in immune/inflammation response processes as the most prominent discriminators between subtoxic/nontoxic exposure and toxic exposure to APAP. The focus in the highest GO categories shifted from general immune response to more specific mechanisms of response with decreasing members of a given gene list (Fig. 2).
Fig. 2.
Differentially expressed genes in blood that discriminate between exposure to subtoxic/nontoxic or toxic dose of APAP. Pictured is a subgroup of genes involved in immune response and inflammation. Gray filling of circles beside genes indicates identification of genes in the list of discriminatory genes (top circle, EPIG; middle circle, k-NN, bottom circle, ANOVA DME and DCE), and coloring of squares signals direction of change (red, up-regulation; green, down-regulation in comparison with mock-treated control).
Pathway analysis of the predictor genes revealed a down-regulation of energy consuming pathways (gluconeogenesis and propionate metabolism) and up-regulation of energy producing pathways (glycogen phosphorylase) after exposure to a toxic dose. Several proapoptotic genes (HLA-DR, PPP1C, and karyopherin-α) were down-regulated, whereas anti-apoptotic Iκ-B was down-regulated after 6 h and up-regulated at later time points.
Discussion
We tested the hypothesis that blood gene expression data can carry information that allows discrimination of levels of certain adverse exposures using a rat model of exposure to subtoxic and toxic doses of APAP. We used three different prediction strategies to determine gene sets as indicators that allow discrimination of subtoxic/nontoxic and toxic dose levels and extracted one to two signature gene sets with each method from the training data set. Those gene sets were tested for prediction accuracy on blinded, independent rat blood and liver test data sets. Although the test data included a time point (3 h) and two dose levels (0 and 2,000 mg/kg APAP) that were not present in the training data set, the signature gene lists were able to predict exposure to subtoxic/nontoxic versus toxic doses with very high accuracy (88.9–95.8%). Prediction of APAP-induced liver injury based on blood gene expression data outperformed predictions based on clinical chemistry, histopathology, and hematology (Table 2). These traditional clinical parameters were particularly inferior (compared with gene expression analysis) at the prediction of exposure levels of animals, which were analyzed at early times after exposure that preceded the development of clinical signs of hepatic injury. When analyzing only the 24-h data in the test set, at which point peak injury had developed, the prediction accuracy of clinical chemistry and histopathology improved, but still did not reach the prediction accuracy of three of the four gene expression predictions.
The analytical methods used for prediction of the blinded samples based on gene expression data used similar data-mining strategies. Each used data reduction and gene selection approaches to narrow down the genes to a candidate set. To ensure the reliability of the results from the analytical procedures used in this pursuit, precautions were taken throughout the study. For instance, in the gene-selection step, a false discovery rate (FDR) was determined to control possible family-wise errors and to balance between type I and type II errors in the statistical models.
Microarray gene expression analysis lends the ability to profile the overall response of thousands of genes simultaneously across several experimental conditions. This approach presents a well known large n (number of genes) and small p (number of samples) problem in statistical analysis. In our study, a small set of genes were obtained and used to build the classifiers for the prediction models. Furthermore, cross-validation procedures were used in the predictions to minimize the overfitting of the classifiers on the training data.
Interestingly, when tested for transorgan prediction accuracy, the EPIG method, yielding the most genes, was also able to predict the toxic dose of exposure when testing expression values from liver samples of the test set animals (after training on blood gene expression data of the training set) with 97.2% accuracy (Table 2 and SI Fig. 5b). The classifier-based methods are more useful for identifying a small, focused set of gene indicators of toxic exposure, whereas pattern-based methods such as EPIG are promising when attempting to identify groups of genes to discern biological processes perturbed by environmental pressures, toxic exposure, or mechanistic changes.
To test whether our predictive gene sets would be useful on human samples, we retrieved human orthologous genes for the union set of the 270 predictor genes and found 66 of them to be present on both the human and rat Agilent chips. Interestingly, cluster analysis using the expression values of these genes from the blood of five human APAP overdose victims and three control individuals (SI Table 9) allowed clear separation dependent on APAP exposure status (SI Fig. 6a). The cosine correlation values from the individual clusters of overdosed victims are high (>+0.9) and moderately high (>+0.8) among them as a whole cluster (SI Fig. 6b). To test the significance of this finding, 10,000 random selections of 66 genes for k-means clustering into two partitions were performed and yielded a probability of 0.0061 that the clustering of the samples would be as stable (10) as the 66 orthologous genes by chance. More data and further analysis are needed to completely understand the significance of these findings for patient samples. However, it is encouraging to see a separation of overdosed and normal individuals based on genes that were retrieved from a rat blood training set.
Analysis of gene expression data, in conjunction with other histopathological and serum parameters, revealed several animals at the 24 h time point in the test set that presented an altered response to the exposure to toxic levels of APAP. In particular, those animals presented no significant elevations of ALT and/or SDH activities in the serum and no or minimal histopathological changes. For example, one animal (no. 3338) in the training set was consistently predicted as a subtoxic-dosed animal and was found to have only minimal necrosis and no ALT or SDH elevation. In the test set, six animals showed this characteristic of having received a toxic dose but showed this altered response (nos. 61, 64, 65, 67, 69, and 70). Of these animals, four were predicted by two or more algorithms as having received subtoxic or nontoxic treatment (SI Table 8). Interestingly, the EPIG analysis grouped five of those six animals in the subtoxic/nontoxic group. This result raises the issue whether those animals would have developed a more pronounced toxic phenotype at a later time point after treatment or whether their response to this level of exposure would never have manifested the severe response seen in the majority of animals. If the latter, then EPIG would qualify as the analysis method that discriminates most accurately based on the actual pathobiology associated with exposure. Because of the study design (endpoint and not longitudinal), we cannot know what the ultimate fate of animals killed at earlier times after exposure would have been, because peak injury is usually reached ≈24 h after exposure in the rat. Histopathological and serum parameters are often not significantly altered before this time point, even after highly toxic exposure levels.
Interestingly, although the three different algorithms used produced signature gene sets of very different numbers and extracted those lists from the data by different approaches, the main biological response captured by each was shared between all of them. An alteration of inflammatory pathways involving interleukin-1 and NF-κB was the main biological difference between exposure to toxic or subtoxic/nontoxic doses of APAP. The role of inflammation in APAP-induced liver injury has been well described (11). The identification of this inflammatory response in a genomic signature in the blood is a previously undescribed finding of our study. This finding might provide one of the missing links for the phenomenon of organ-to-organ communication seen after APAP-induced toxicity as described by Neff et al. (12). These authors described alterations of cytokine/chemokine expression in the liver in response to APAP exposure that affect the lung. Our results may have three interpretations. First, the blood might react in the same manner as the lung to the release of cytokines and chemokines by the liver, with the inflammatory patterns we observed in response to those stimulants. Second, the inflammatory response we observed might be a direct result of blood cells being exposed to APAP and its toxic metabolites and occurring parallel to a similar response in the liver. Third, APAP could be producing a general response in the blood common to other agents that induce liver injury or a nonliver inflammatory (immune) response. These possibilities are not mutually exclusive and several might occur simultaneously. By using gene expression data acquired from the blood of rats exposed to a compendium of hepatotoxicants (13), some of which elicit liver injury (centrilobular necrosis) similar to APAP, as well as data from a study in which rats were treated with the inflamogen LPS (14), our 270 genes clearly show a different pattern of expression of APAP-toxic exposure in comparison with the compendium of hepatotoxicants, and they also distinguish the LPS-treated animals from those exposed to APAP (SI Fig. 7). For example, N-nitrosomorpholine at 300 mg/kg exposure to the rat liver for 48 h causes elevations in liver injury enzyme markers aspartate aminotransferase (AST) and ALT, and centrilobular necrosis is manifested to a marked severity level (see SI Table 10). However, the cluster analysis of the 270 genes' expression data from the APAP-treated samples and other hepatotoxicants shows that the samples exposed to N-nitrosomorpholine cluster adjacent to the APAP-toxic samples, but there are clear differences in the expression profiles of some of the genes. Furthermore, the pattern of our 270 genes did not group the LPS animals with either the toxic or subtoxic APAP doses.
We conclude that blood gene expression data can provide signatures that are good predictors of exposure to toxic doses of APAP that are superior to traditional toxicological parameters, especially at early time points after exposure. A diagnostic test that would help in the identification and prognosis of individuals with APAP-induced hepatotoxicity would be clinically useful. It will be intriguing to further test to what extent our result can be translated into the clinical setting with individuals presenting with APAP intoxication.
Materials and Methods
Animals and Animal Care.
Male F344/N rats, 10–12 weeks old, were obtained from Taconic Laboratories (Germantown, NY) and provided with NIH-07 diet and tap water ad libitum.
Chemical.
APAP (99% pure) was purchased from Sigma (St. Louis, MO), and suspension formulations were prepared by mixing with 0.5% aqueous ethyl cellulose (USP/FCC grade; Fisher Scientific, St. Louis, MO).
Study Design.
For the training set, groups of four male rats, 12–14 weeks old, not fasted before dosing, each received 0 (vehicle only), 150, 1,500 or 2,500 mg/kg APAP in 0.5% ethyl cellulose by oral gavage in two doses to increase absorption. The animals were killed after 6, 12, or 24 h. For the test set, groups of six male rats each received 0 (vehicle only), 150, 1,500, or 2,000 mg/kg APAP in 0.5% ethyl cellulose by oral gavage in two doses. These animals were killed after 3, 6, or 24 h. An earlier time point and a lower toxic dose were incorporated in the test data set to ensure that the predictive power is not limited to the exact conditions represented by the training set. Experiments were performed according to the guidelines established in the National Institutes of Health Guide for the Care and Use of Laboratory Animals (15), and an approved Animal Study Protocol was on file before initiation of the study.
Histopathology.
A study pathologist initially evaluated two H&E-stained sections of left liver lobes. A second pathologist reviewed the diagnosis. Discrepancies between the pathologists were resolved by a pathology working group review (16).
For the blinded histopathology evaluation of the test set, three pathologists evaluated the training set. They received the left liver lobe histopathology slides of the test set in a blinded fashion and were charged to group the slides based on the criteria that (i) no pathology is seen or (ii) pathology is seen that indicates hepatotoxic insult (hepatocyte degeneration and necrosis) to the animal.
Clinical Pathology.
Blood was collected at euthanization into serum separation tubes (BD Microtainer tubes; BD, Franklin Lakes, NJ), and serum was separated. Clinical chemistry analyses (albumin, cholesterol, creatinine, direct bilirubin, total bilirubin, total bile acid concentrations, triglycerides, and activities of ALT, alkaline phosphatase, aspartate aminotransferase, lactate dehydrogenase, and SDH) were performed on all rats at study termination.
Hematology.
Blood was collected in EDTA tubes (BD Microtainer tubes). Complete blood counts (white blood cells, red blood cells, hemoglobin, hematocrit, and platelets), reticulocyte counts and differential white blood cell counts (neutrophils, lymphocytes, monocytes, eosinophils, and basophils) were performed on all rats at study termination.
RNA Isolation.
At euthanization, two liver sections from the left lobe were isolated for histopathology evaluation and fixed in 10% formalin. The remaining tissue of the left liver lobe was cubed, flash frozen, and pulverized as described (17). Total hepatic RNA was isolated from individual rat livers with Qiagen RNeasy Maxi Kits (Qiagen, Valencia, CA) as described (18). Blood was collected by using PAXgene vacutainer tubes (PreAnalytiX; Qiagen), and RNA was isolated as described (14).
For the training set, equal amounts of blood RNA from each of four vehicle-only-treated control animals at the 6 and 12 h time points and from each of six vehicle-only-treated control animals at the 24 h time point were pooled for control gene expression. These pools were compared with individual treated animals at each dose and time period. For the test set, equal amounts of blood or liver RNA from each of six vehicle-only-treated control animals were pooled at each time point and compared with individual control and treated animals at each dose and time point. The samples were hybridized in duplicate (fluor-flips) for each individual rat. Thus, for each dose and time period, 8 arrays were performed for the training set (with the exception of only 6 for both the 150 mg/kg APAP and 1,500 mg/kg dose groups) and 12 arrays for the test set.
Microarray Analysis.
RNA samples were labeled with Cy3 and Cy5 with the Agilent Fluorescent Linear Amplification Kit (Agilent, Palo Alto, CA) and hybridized to Agilent Rat Oligonucleotide Microarrays (Agilent G4130A) according to the manufacturer's instructions. Fluorescent intensities were measured with an Agilent DNA Microarray Scanner (Agilent G2565AA) and processed with the Agilent G2565AA Feature Extraction Software. Detailed protocols are available at www.niehs.nih.gov/research/atniehs/core/microarrays. The complete data set has been deposited at the Gene Expression Omnibus (GEO) Database under accession no. GSE5652.
Identification of Significantly Expressed Genes.
The gene expression data were loaded into the Rosetta Resolver database (build 5.1.0.1.23, Rosetta Inpharmatics; Agilent Technologies, Palo Alto, CA) and merged according to fluor-flip hybridization pairs to generate weighted-averaged ratio values (computed from the normalized and background-subtracted pixel intensity values). An error-weighted (19, 20) two-way ANOVA model was constructed with the gene expression data to capture both dose and time main effects, as well as the dose/time interaction for a comparison among doses (150, 1,500, and 2,500 mg/kg) of the training samples
where y is the log base 10 of the ratio value for the kth gene, μ represents the grand mean, T is the main effect for time, D is the main effect for dose, (TD)ij is the interaction of ith time and jth dose, and ε is the term for stochastic error. An error-weighted one-way ANOVA was also constructed with the gene expression data to capture the dose effect confounded by time of exposure.
Genes identified as significantly expressed between subtoxic-dosed (150 mg/kg) and toxic-dosed (1,500 and 2,500 mg/kg) training set samples were used for gene selection. Missing data had their log base 10 value replaced with 0.
ANOVA Gene Selection and k-NN Classifier Building.
For gene selection, a one-way ANOVA on the two classes of the training data (subtoxic dose, 150 mg/kg; toxic dose, 1,500 and 2,500 mg/kg) was constructed to identify the genes that had the highest significance of the difference between the two classes. To build the classifier, the k-NN approach was implemented by using Euclidean distance and k = 3. The k-NN classifier model was constructed by using the training data, and its accuracy was validated with a 10-fold cross-validation scheme. This type of validation procedure has been proven to have a low mean square error and small bias of the classifier (21). A 10-fold cross-validation approach was taken to render the highest assessment of classification precision without extra computational cost or loss of classifier generalization. The classifier was built by using Partek Pro 6.0 (build 6.04.1112; Partek, St. Louis, MO).
SVM for Gene Selection.
The Gene Expression Model Selector software (22) was used to construct an MC-SVM from a linear kernel for gene selection. Two classes from the training data were generated: a subtoxic-dosed class containing the samples treated with 150 mg/kg APAP and a toxic-dosed class containing samples treated with 1,500 mg/kg or 2,500 mg/kg APAP. Briefly, using the training data, two binary SVM classifiers were constructed with the subtoxic class versus the toxic class. A minimum of 20 genes for selection as predictors were optimized by using signal-to-noise ratio in a one-versus-rest comparison approach (stepped by including five genes at each iteration of the algorithm). Validation of the predictor genes was performed by using LOOCV. Accuracy was determined by the proportion of correct classifications over the total number of classifications.
Simplified Fuzzy ARTMAP Prediction.
The genes selected as highly accurate predictors of the class of the samples in the training set via the MC-SVM were used to compile the ratio values from the test data sets. The simplified Fuzzy ARTMAP tracking neural network (7, 8) was used to build a classifier and for predicting the class of the samples in the test sets. Methodology and the software for performing the prediction are available at www.niehs.nih.gov/research/resources/software/exp. A vigilance parameter of 0.2 was one of the other values to give maximal accuracy of the classifier and was used during prediction of the APAP test sets.
Pattern Extraction and Gene Selection for Prediction.
The extraction of gene expression patterns and identification of genes (via EPIG) that distinguished between subtoxic-dosed and toxic-dosed samples was performed. Briefly, ratio intensity values from all of the genes on the arrays were log base 2 transformed, adjusted by systematic variation normalization (23) and corrected for dye bias. An outlier animal (no. 3338) was not included in the analysis. A set of nine intra-groups (samples at the three time points for each of the three given doses) were formed. Distinct patterns of gene expression were extracted based on the expression profiles correlation values, the minimum cluster size for the patterns, and the cluster-partitioning resolution (9). From the patterns and using signal-to-noise ratio = 3, magnitude = 1.5, and a correlation r value of 0.64 of the gene profiles, genes were selected and included in the signature list for prediction. PCA of the training and the test samples simultaneously was performed to predict the classes of the samples by visualizing the closeness of the test samples to either class of the training samples.
Biological Pathway Analysis.
For each predictive gene list and for the union of all four lists, GO analysis was performed in Rosetta Resolver. Genes involved in the top categories were selected and analyzed in the Ingenuity Pathway Analysis tool (Ingenuity Systems, Redwood City, CA). Additionally, the complete predictive gene lists from all of the prediction methods and the union of those genes were analyzed with the Ingenuity Pathway Analysis tool and with Metacore (GeneGo, St. Joseph, MI).
Human Blood Analysis.
Blood was drawn from normal healthy volunteers or APAP-overdose patients admitted to the University of North Carolina Emergency Room under UNC IRB protocol 04-MED-416 (SI Table 11). RNA was purified by using the PAXgene system as above and analyzed by using Agilent Human Oligonucleotide 1Av2 microarrays.
Supplementary Material
Acknowledgments
We thank Pamela Blackshear for excellent support with histopathological evaluations; Edward Lobenhofer, Todd Auman, and Gail Carpenter for stimulating discussions that supported the preparation of this manuscript; Ben Van Houten, Steven Kleeberger, and Douglas Bell for critical review of the manuscript, and Boston University for granting the authors permission to use Fuzzy ARTMAP. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH) and the National Institute of Environmental Health Sciences (NIEHS). This work also was funded in part with Federal funds from NIEHS, NIH, under Contracts N01-ES-25497, N01-ES-95442, and NO1-ES-35513.
Abbreviations
- APAP
acetaminophen
- DME
dose main effect
- DCE
dose confounded effect
- MC-SVM
multicategory support vector machines
- EPIG
extracting patterns and identifying genes
- k-NN
k-nearest neighbors
- LOOCV
leave-one-out cross validation
- PCA
principal component analysis
- GO
Gene Ontology
- Fuzzy ARTMAP
fuzzy adaptive resonance theory map
- ALT
alanine aminotransferase
- SDH
sorbitol dehydrogenase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data have been deposited in the Gene Expression Omnibus (GEO) Database, www.ncbi.nlm.nih.gov/geo (accession no. GSE5652).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706987104/DC1.
References
- 1.Larrey D, Pageaux GP. Eur J Gastroenterol Hepatol. 2005;17:141–143. doi: 10.1097/00042737-200502000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Watkins PB, Kaplowitz N, Slattery JT, Colonese CR, Colucci SV, Stewart PW, Harris SC. J Am Med Assoc. 2006;296:87–93. doi: 10.1001/jama.296.1.87. [DOI] [PubMed] [Google Scholar]
- 3.Tsai CL, Chang WT, Weng TI, Fang CC, Walson PD. Clin Ther. 2005;27:336–341. doi: 10.1016/j.clinthera.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Blei AT. Liver Transpl. 2005;11:S30–4. doi: 10.1002/lt.20595. [DOI] [PubMed] [Google Scholar]
- 5.Dinkel HP, Wittchen K, Hoppe H, Dufour JF, Zimmermann A, Triller J. Rofo. 2003;175:1112–1119. doi: 10.1055/s-2003-40910. [DOI] [PubMed] [Google Scholar]
- 6.Terjung B, Lemnitzer I, Dumoulin FL, Effenberger W, Brackmann HH, Sauerbruch T, Spengler U. Digestion. 2003;67:138–145. doi: 10.1159/000071293. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter G, Grossberg S, Markuzon N, Reynolds JH, Rosen DB. IEEE Trans Neural Netw. 1992;3:698–713. doi: 10.1109/72.159059. [DOI] [PubMed] [Google Scholar]
- 8.Kasuba T. AI Expert. 1993;8:18–25. [Google Scholar]
- 9.Zhou T, Chou JW, Simpson DA, Zhou Y, Mullen TE, Medeiros M, Bushel PR, Paules RS, Yang X, Hurban P, et al. Environ Health Perspect. 2006;114:553–559. doi: 10.1289/ehp.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Famili AF, Liu G, Liu Z. Bioinformatics. 2004;20:1535–1545. doi: 10.1093/bioinformatics/bth124. [DOI] [PubMed] [Google Scholar]
- 11.Luster MI, Simeonova PP, Gallucci RM, Bruccoleri A, Blazka ME, Yucesoy B. Toxicol Lett. 2001;120:317–321. doi: 10.1016/s0378-4274(01)00284-3. [DOI] [PubMed] [Google Scholar]
- 12.Neff SB, Neff TA, Kunkel SL, Hogaboam CM. Exp Mol Pathol. 2003;75:187–193. doi: 10.1016/s0014-4800(03)00096-0. [DOI] [PubMed] [Google Scholar]
- 13.Lobenhofer EK, Boorman GA, Phillips KL, Heinloth AN, Malarkey DE, Blackshear PE, Houle C, Hurban P. Toxicol Pathol. 2006;34:921–928. doi: 10.1080/01926230601072319. [DOI] [PubMed] [Google Scholar]
- 14.Fannin RD, Auman JT, Bruno ME, Sieber SO, Ward SM, Tucker CJ, Merrick BA, Paules RS. Physiol Genomics. 2005;21:92–104. doi: 10.1152/physiolgenomics.00190.2004. [DOI] [PubMed] [Google Scholar]
- 15.Council NR. Guide for the Care and Use of Laboratory Animals. Washington, DC: Natl Acad Press; 1996. [Google Scholar]
- 16.Boorman GA, Eustis SL. Managing Conduct and Data Quality of Toxicological Studies. Princeton, NJ: Princeton Sci Pub; 1986. [Google Scholar]
- 17.Foley JF, Collins JB, Umbach DM, Grissom SF, Boorman GA, Heinloth AN. Toxicol Pathol. 2006;34:795–801. doi: 10.1080/01926230601009527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamadeh HK, Knight BL, Haugen AC, Sieber S, Amin RP, Bushel PR, Stoll R, Blanchard K, Jayadev S, Tennant RW, et al. Toxicol Pathol. 2002;30:470–482. doi: 10.1080/01926230290105712. [DOI] [PubMed] [Google Scholar]
- 19.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 20.Stoughton R, Dai H. US Patent 6. 2002;351:712. [Google Scholar]
- 21.Molinaro AM, Simon R, Pfeiffer RM. Bioinformatics. 2005;21:3301–3307. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- 22.Statnikov A, Aliferis CF, Tsamardinos I, Hardin D, Levy S. Bioinformatics. 2005;21:631–643. doi: 10.1093/bioinformatics/bti033. [DOI] [PubMed] [Google Scholar]
- 23.Chou JW, Paules RS, Bushel PR. J Bioinform Comput Biol. 2005;3:225–241. doi: 10.1142/s0219720005001028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.