Abstract
Influenza viruses infect vertebrates, including mammals and birds. Influenza virus reverse-genetics systems facilitate the study of the structure and function of viral factors. In contrast, less is known about host factors involved in the replication process. Here, we developed a replication and transcription system of the negative-strand RNA genome of the influenza virus in Saccharomyces cerevisiae, which depends on viral RNAs, viral RNA polymerases, and nucleoprotein (NP). Disruption of SUB2 encoding an orthologue of human RAF-2p48/UAP56, a previously identified viral RNA synthesis stimulatory host factor, resulted in reduction of the viral RNA synthesis rate. Using a genome-wide set of yeast single-gene deletion strains, we found several host factor candidates affecting viral RNA synthesis. We found that among them, Tat-SF1, a mammalian homologue of yeast CUS2, was a stimulatory host factor in influenza virus RNA synthesis. Tat-SF1 interacted with free NP, but not with NP associated with RNA, and facilitated formation of RNA-NP complexes. These results suggest that Tat-SF1 may function as a molecular chaperone for NP, as does RAF-2p48/UAP56. This system has proven useful for further studies on the mechanism of influenza virus genome replication and transcription.
Keywords: molecular chaperone, replication, RNA-dependent RNA polymerase, nucleoprotein
Viruses are intracellular parasites. Virus replication requires virus-derived factors and also depends totally on host cell functions/machinery. Identification of host factors involved in viral replication is critical for understanding the molecular mechanism of virus replication and pathogenicity. In the case of DNA viruses, a number of host factors are identified; their functional analyses not only have contributed to understanding the molecular mechanism of viral genome replication and transcription but also have led to the study of eukaryote genome replication and transcription (1). The RNA genome of positive-strand RNA viruses is “infectious,” where infectious means the infectious virus can be recovered when the RNA genome is introduced into cells (2–4). Thus, reverse-genetic systems using in vitro synthesized RNAs of the positive-strand RNA virus were powerful to reveal the role of viral factors and the interaction between viral and host factors (5). In contrast, for the generation of an infectious negative-strand RNA virus, the negative-strand virus RNA genome should be introduced into cells as complexes, with viral RNA polymerases and other viral factors required for RNA-dependent RNA synthesis. Alternatively, the negative-strand RNA genome should be introduced into cells expressing these viral factors.
The influenza virus contains segmented- and negative-strand RNAs as its genome. Influenza virus RNA is associated with viral RNA-dependent RNA polymerases consisting of PB1, PB2, and PA subunits and nucleoprotein (NP)-forming viral ribonucleoprotein complexes (vRNP) (6). vRNP is a basic unit for transcription and replication of the virus genome. It was shown that vRNP complexes isolated from virions are “infectious” (7). Then, transfection systems were established using reconstituted vRNP complexes from which genome replication and transcription proceed (8, 9). Recently, a reverse-genetics system was established for the generation of a recombinant influenza A virus from a set of plasmids (10). With this system, the structure and function of viral factors have been studied extensively (11, 12).
Recent proteomics have shown a list of cellular proteins that interact with viral proteins (13). However, only a few host factors have been identified by functional assays for viral genome transcription and replication (14–19). Further, a systematic screening system has been needed to identify host factors. Yeast is a good model eukaryotic cell, with merits including well established genetics and information on the entire genome for genome-wide screening. It has been shown that yeast cells support the replication and transcription of some positive-strand viral RNA genomes such as brome mosaic virus and tomato bushy stunt virus (20, 21).
In this study, to identify host factors systematically, we tried to develop a system in which yeast cells support the replication and transcription of the influenza virus genome depending on transfected vRNP complexes. With this system, we confirmed that the yeast orthologue of a previously identified mammalian host factor is indeed a stimulatory factor for viral RNA synthesis in yeast cells. In addition, we identified host factor candidates for the regulation of virus RNA synthesis using a yeast single-gene knockout library. Among these candidates, Tat stimulatory factor 1 (Tat-SF1), a mammalian homologue of a newly identified candidate, CUS2, was a stimulatory host factor in influenza virus RNA synthesis. Thus, this system could be quite useful for understanding the molecular mechanism of virus replication and could provide a method for systematic screening of host factors in the influenza virus genome replication process.
Results
Replication and Transcription of the Influenza Virus Genome in vRNP-Transfected Yeast Cells.
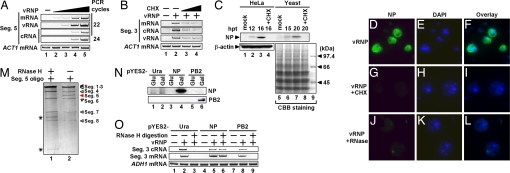
First, to examine whether vRNP purified from virions is “infectious” in yeast cells, we introduced vRNP into the cells. The synthesis level of viral RNAs, i.e., vRNA, cRNA (complementary RNA; the template for amplification of vRNA), and viral mRNA derived from segment 5 vRNA were analyzed by RT-PCR (Fig. 1A). The amount of viral RNAs synthesized in vRNP-transfected yeast cells increased depending on increasing amounts of transfected vRNP. The amount of viral mRNA and vRNA synthesized in yeast cells was more than that of cRNA (Fig. 1A; see the PCR result with 22 cycles), as reported using infected mammalian cells (12, 22). A similar result was observed for other viral segments (data not shown). These results demonstrate that viral RNA polymerase and NP are functional in yeast cells. In infected mammalian cells, primary viral transcription depends on infecting vRNP, whereas viral genome replication requires newly synthesized viral proteins (12). When vRNP-transfected yeast cells were treated with cycloheximide (CHX), a protein synthesis inhibitor, the synthesis level of viral RNAs was markedly reduced (Fig. 1B), indicating that the synthesis of viral RNAs depends on newly synthesized proteins. Because viral mRNA synthesis was also sensitive to CHX treatment, viral mRNA could be synthesized from newly synthesized vRNA as a template in this system (12, 22). By Western blot analyses [Fig. 1C; see supporting information (SI) Text for detailed discussion], we detected viral proteins synthesized in vRNP-transfected yeast cells. In indirect immunofluorescence assays using anti-NP antibody (Fig. 1 D–L), NP was detected in vRNP-transfected yeast cells (Fig. 1D) but not in cells treated with CHX (Fig. 1G). When vRNP was treated with RNase A before transfection, the expression of NP was abolished (Fig. 1J), indicating that viral gene expression depended on vRNA in vRNP complexes. Taken together, we conclude that yeast cells support viral genome replication and viral gene transcription depending on transfected vRNP complexes.
Fig. 1.
Replication and transcription of the influenza virus genome in yeast cells. (A) Yeast spheroplasts were mock-transfected (lane 1) or transfected with vRNP (0.1, 0.3, 1, and 2 μg of NP equivalents for lanes 2, 3, 4, and 5, respectively) purified from virions. At 48 h posttransfection (hpt), total yeast RNA was extracted and subjected to reverse transcription. PCR was then performed with primer sets specific for negative- (vRNA) and positive-sense RNA (mRNA or cRNA) of segment 5 and ACT1 mRNA. Amplified double-stranded DNAs were subjected to 7% PAGE and visualized by ethidium bromide. (B) Yeast cells transfected with vRNP were incubated in the absence (lanes 1 and 2) or presence of 3 mg/ml (lane 3) and 10 mg/ml (lane 4) cycloheximide (CHX). RT-PCR was performed with primers specific for segment 3 RNA and ACT1 mRNA. (C) Viral protein synthesis in vRNP-transfected yeast and mammalian cells. HeLa cells or yeast cells were transfected with 3 μl of RNP (450 ng of NP equivalents). After transfection, cells were incubated in the absence (lanes 1–3 and 5–7) or presence of 100 μg/ml (lane 4) and 3 mg/ml (lane 8) CHX. We used the previously described method (40) for the preparation of total protein from yeast cells. HeLa cell (20 μg) or yeast cell lysates (22.5 μg) were loaded for each lane. Western blot analyses were carried with anti-NP or -β-actin antibodies as a control. Coomassie brilliant blue (CBB) staining is also shown as a control for the preparation of yeast lysates because of the lack of an appropriate antibody. The molecular mass marker is shown in lane 9. (D–L) vRNP-transfected yeast cells were immunostained at 24 hpt. NP and DNA were stained with anti-NP antibody (D, G, and J) and DAPI (E, H, and K), respectively. Overlay of NP and DAPI staining panels (F, I, and L). Yeast cells were incubated in the absence (D–F and J–L) or presence (G–I) of 5 mg/ml CHX. Before transfection, vRNPs were treated with RNase A for 10 min at 37°C (J–L). (M–O) The complementation experiment of segment 5-depleted vRNP. (M) Digestion of segment 5 vRNA. The vRNP (3 μg of NP equivalents) was mixed with 300 ng of an oligonucleotide corresponding to part of segment 5 vRNA (segment 5 digestion; see SI Text) in the presence of 0.4 M NaCl for 5 min at 37°C and then treated (lane 1) or mock-treated (lane 2) with 30 units of RNase H for 5 min at 37°C. Purified RNA was loaded onto a 3.2% polyacrylamide gel containing 7.7 M urea and visualized by silver staining. Asterisks indicate bands possibly corresponding to digested fragments. (N) Western blot analysis of induced viral proteins. Control (lanes 1 and 2), NP (lanes 3 and 4), and PB2 (lanes 5 and 6). (O) RT-PCR analysis of viral RNAs. vRNP and vRNP devoid of segment 5 vRNA (RNase H digestion) were transfected into yeast cells transformed with pYES2 (lanes 1–3), pYES2-NP (lanes 4–6), and pYES2-PB2 (lanes 7–9). Yeast cells were incubated for 24 h in medium containing galactose. RT-PCR analysis was performed with primer sets specific for segment 3 cRNA, mRNA, and ADH1 mRNA.
We performed complementation experiments using yeast cells transfected with vRNP complexes devoid of segment 5 vRNA that encodes NP. NP is required for formation of vRNP complexes and efficient elongation of the viral RNA chain (6). We eliminated segment 5 vRNA in vRNP complexes by digestion with RNase H (Fig. 1M) (see SI Text) (23). Then, vRNP complexes lacking segment 5 vRNA (designated depleted vRNP) and mock-digested vRNP complexes were introduced into yeast cells expressing NP or PB2. Fig. 1N confirmed the expression level of NP and PB2 induced by galactose using pYES2-NP and pYES2-PB2, in which NP and PB2 genes are under the control of the GAL1 promoter (see SI Text). In yeast cells transformed with pYES2 or pYES2-PB2, depleted vRNP did not lead to the synthesis of mRNA and cRNA from segment 3 vRNA (Fig. 1O, lanes 3 and 9). In contrast, yeast cells transformed with pYES2-NP could rescue RNA synthesis (Fig. 1O, lane 6) in the galactose induction medium. These results indicate that exogenously added NP complements the system using depleted vRNP. Further, it is now shown that the replication process in yeast cells depends on NP.
Effect of sub2 Deletion.
We tried to use the system for the functional analysis of RAF-2p48, a previously identified host factor (14). RAF-2p48 facilitates formation of NP-RNA complexes and stimulates in vitro RNA synthesis from a model viral RNA. RAF-2p48 is identical to UAP56, an RNA splicing factor. SUB2, a putative Saccharomyces cerevisiae orthologue of human RAF-2p48/UAP56 (62% amino acid sequence identity), associates with Sm snRNPs and is involved in splicing in vivo (24, 25). Before genetic analysis of sub2 in yeast cells, we confirmed that SUB2 stimulates influenza virus RNA synthesis in vitro (SI Fig. 5A). The viral RNA synthesis stimulatory activity of SUB2 was as much as 50% of RAF-2p48/UAP56. This result suggests that SUB2 may function as a host factor for viral genome replication in yeast cells. It is possible that not only conserved but also diverse regions between SUB2 and UAP56 are required for the full activity of RAF-2p48. Then, we examined the effect of the deletion of sub2 on viral RNA synthesis in yeast cells. The sub2 deletion strain was constructed by replacing its ORF with a TRP1 maker (see SI Text). We analyzed the level of viral RNA synthesis in the sub2 deletion yeast strain by real-time RT-PCR. In the vRNP-transfected sub2 deletion strain, the rate of cRNA synthesis was reduced to 40% that in a wild-type strain at 9 h after transfection (SI Fig. 5B). This suggests that SUB2 facilitates viral RNA synthesis as a host factor in yeast cells, and thereby that RAF-2p48/UAP56 is a host factor for viral RNA synthesis in vertebrate cells. Viral RNA synthesis in the sub2 deletion strain rescued up to 60% of that in the wild-type strain after longer incubation, so we assume yeast cells have another host factor(s) that complements the function of SUB2.
Identification of Host Factor Candidates Affecting Viral Genome Replication.
This yeast system could allow us to identify more host factor candidates. Once we reached a candidate, we went back to the vertebrate system to verify and characterize the candidate. To this end, we carried out screening of a yeast single-gene deletion library containing ≈4,800 strains (80% of all yeast genes) (26). We first focused on screening deletion strains lacking 354 genes encoding putative nucleic acid-binding/-related functional proteins among nuclear genes [categorized by advanced search of the Saccharomyces Genome Database (www.yeastgenome.org)]. Each deletion strain was transfected with vRNP, and the synthesis level of segment 7 cRNA was analyzed by RT-RCR (data not shown). In some deletion strains, such as those lacking lsm12, npl3, etc., viral RNA synthesis was increased compared with the wild-type strain. These deleted gene products are candidates for inhibitory factors for viral RNA synthesis. On the other hand, several strains (mip6, nsr1, etc.) showed a severe decreased level of viral RNA synthesis. Interestingly, the cRNA synthesis rate was decreased in the deletion strains of several factors involved in splicing. The synthesis level of segment 3 cRNA was quantitatively determined by real-time RT-PCR in mutants containing deletions in genes encoding splicing factors (SI Fig. 6). The significant decreased phenotype was found in strains lacking three genes, isy1, msl1, and cus2. The human homologue of the ISY1 gene (KIAA1160 cDNA) (27) encodes an unidentified protein. The human MSL1 homologue is a U1 snRNP-specific protein A (U1-A) associated with the 164-nt-long U1 snRNA (28). CUS2 shares 37% identity with human Tat-SF1 (29), a transcription elongation protein (30–32). Tat-SF1-snRNP complexes are recruited to the cellular RNA polymerase II elongation complex through the binding of Tat-SF1 and snRNPs to p-TEFb and the nascent splicing substrate, respectively (33).
Tat-SF1 Functions as a Host Factor in Infected Cells.
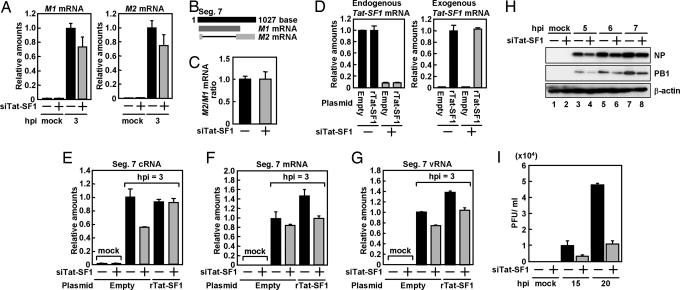
Next, we attempted to characterize the function of Tat-SF1 in influenza virus genome replication. To examine whether Tat-SF1 is involved in viral RNA synthesis in mammalian cells, we carried out knockdown experiments of Tat-SF1 with the siRNA technique. Using the cells in which the amount of Tat-SF1 mRNA was reduced (SI Fig. 7A), we determined the synthesis level of segment 7 viral RNAs. The synthesis level of viral cRNA in Tat-SF1 knockdown cells (SI Fig. 7B, lane 4) was <10% of that in cells transfected with control plasmids (SI Fig. 7B, lanes 1 and 3; mock and 6 h after infection). These results suggest that Tat-SF1 plays a role in the viral RNA synthesis, although it is possible that the decreased level of Tat-SF1 may have some effect on splicing viral and host pre-mRNAs and thereby on viral RNA synthesis. To rule out this possibility, we compared the ratio of the level of M1 mRNA to that of M2 mRNA generated from M1 mRNA by splicing both control and Tat-SF1 siRNA (siTat-SF1)-transfected cells. The segment 7 vRNA codes for M1 and M2 proteins are translated from nonspliced and spliced mRNA, respectively (Fig. 2B). In siTat-SF1-transfected cells, the amount of M1 and M2 mRNAs was reduced to 75% of that in mock-transfected cells (Fig. 2A), whereas the ratio of the M1 to the M2 mRNA level in Tat-SF1 knockdown cells was similar to that in control cells (Fig. 2C). Next, we carried out complementation experiments for siRNA-introduced cells with a plasmid encoding FLAG-Tat-SF1 containing a silent mutation within the siRNA target sequence (rTat-SF1) (Fig. 2D). Transfection of the siTat-SF1 led to 40% reduction in segment 7 cRNA synthesis in infected cells compared with the level in cells transfected with the control siRNA (Fig. 3E), whereas the cRNA synthesis level was rescued by transfection of FLAG-rTat-SF1 expression plasmid in siTat-SF1-transfected cells. Similar results were obtained in segment 7 mRNA and vRNA synthesis (Fig. 2 F and G). This was also the case for viral RNA synthesis derived from segment 3 (SI Fig. 8 A–C and see SI Text for details).
Fig. 2.
Effect of Tat-SF1 knockdown on virus infection. (A) Total RNA was prepared from HeLa cells transfected with control siRNA or siTat-SF1 and superinfected with influenza virus. Real-time RT-PCR was carried out with primer sets specific for M1 mRNA, M2 mRNA, and β-actin mRNA. (B) Representation of M1 and M2 mRNA generated from segment 7. (C) The ratio of the amount of M2 mRNA to that of M1 mRNA in HeLa cells transfected with control siRNA or siTat-SF1 and those infected with influenza virus. (D–G) HeLa cells were transfected with control siRNA or siTat-SF1. At 72 hpt, cells were transfected with pCAGGS-FLAG-rTat-SF1 and pCAGGS-empty plasmids. After 24-h incubation, cells were superinfected with influenza virus. Total RNA was prepared from cells at 3 h postinfection (hpi). Real-time RT-PCR was carried out with primer sets specific for endogenous Tat-SF1 (D Left), exogenous FLAG-rTat-SF1 (D Right), segment 7 RNAs (E, cRNA; F, mRNA; G, vRNA), and β-actin mRNA. The results are normalized as the ratio to the level of β-actin mRNA. Error bars show standard deviation. (H) Western blot analyses of viral proteins. Mock- (lanes 1, 3, 5, and 7) or siTat-SF1 siRNA-transfected (lanes 2, 4, 6, and 8) HeLa cells were infected with influenza virus at an moi of 5. Western blot analyses were carried with anti-NP, -PB1, or -β-actin antibodies. (I) Single-step virus growth. Mock- or siTat-SF1 siRNA-transfected HeLa cells were infected with influenza virus at an moi of 0.1. Virus titer was examined by plaque assay at the indicated times after infection.
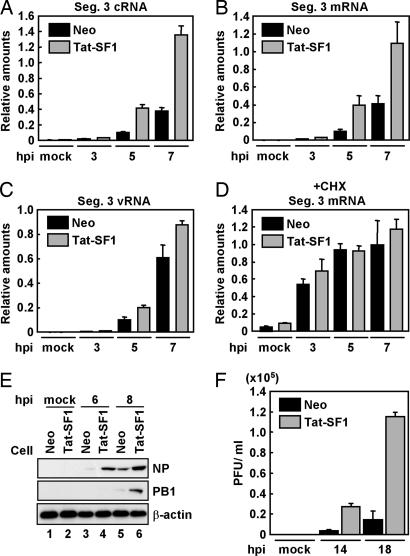
Fig. 3.
Tat-SF1 as a stimulatory host factor involved in virus RNA synthesis. (A–D) Viral RNA synthesis in the presence of a protein synthesis inhibitor. HeLa cells expressing FLAG-Tat-SF1 (Tat-SF1) or control (Neo) HeLa cells were infected with influenza virus in the absence (A–C) or presence (D) of 100 μg/ml CHX. Real-time RT-PCR was carried out with primer sets for segment 3 cRNA (A), segment 3 mRNA (B and D), segment 3 vRNA (C), and β-actin mRNA. Error bars show standard deviation. (E) Western blot analyses of viral proteins. HeLa cells expressing FLAG-Tat-SF1 (lanes 2, 4, and 6) or control HeLa cells (lanes 1, 3, and 5) were infected with influenza virus at an moi of 1. Western blot analyses were carried out with anti-NP, -PB1, or -β-actin antibodies. (F) Single-step virus growth. HeLa cells expressing FLAG-Tat-SF1 or control HeLa cells were infected with influenza virus at an moi of 0.1. Virus titer was examined as described.
We also examined viral protein synthesis and the production of infectious influenza virus in siTat-SF1-transfected cells. The expression of NP and PB1 was reduced in siTat-SF1-transfected cells (Fig. 2H); the virus titer was examined by plaque assay (Fig. 2I). In siTat-SF1-transfected cells, the level of infectious progeny viruses was reduced to 25% of that in control cells. Progeny viruses were not recovered from infected control and siTat-SF1-transfected cells in the presence of CHX (data not shown).
Tat-SF1 Stimulates Viral RNA Synthesis.
To further examine the mechanism of the Tat-SF1-dependent stimulation of viral RNA synthesis, we established a HeLa cell line overexpressing FLAG-Tat-SF1. In cells expressing FLAG-Tat-SF1, viral cRNA, mRNA, and vRNA synthesis from segment 3 was increased (Fig. 3 A–C). This was also the case for viral RNA synthesis derived from segment 7 (SI Fig. 9 A–C and see SI Text for further discussion). It was indicated that CHX suppresses viral protein synthesis and blocks vRNA replication (12), whereas the level of primary transcription from incoming vRNP is not affected in the presence of CHX. Fig. 3D shows that primary transcription in the presence of CHX is not affected by the expression of FLAG-Tat-SF1. These results suggest that Tat-SF1 stimulates the viral RNA synthesis reaction, including replication processes after primary transcription. Thus, it is possible that the enhancement of the viral mRNA synthesis in the absence of CHX (Fig. 3B) is due to the amplified genome by replication.
The effect of the overexpression of FLAG-Tat-SF1 on viral protein synthesis and on the production of infectious progeny viruses was investigated. NP and PB1 were synthesized at the trace level in control cells at 6 and 8 h, respectively, after infection, whereas the synthesis of NP and PB1 was markedly increased in cells expressing FLAG-Tat-SF1 (Fig. 3E). Further, overexpression of Tat-SF1 led to a 5- to 6-fold increase in progeny virus production compared with control cells (Fig. 3F).
Tat-SF1 Facilitates the Formation of the vRNA-NP Complex.
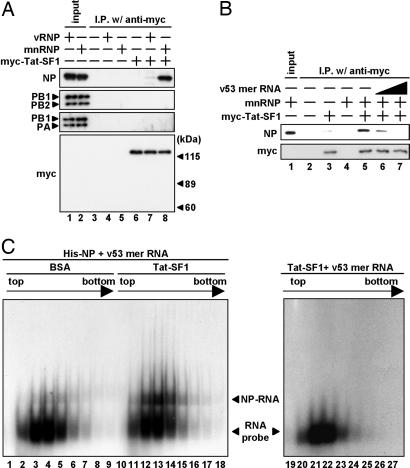
To determine a viral factor(s) that interacts with Tat-SF1, we carried out immunoprecipitation assays using recombinant His-myc-Tat-SF1 (designated myc-Tat-SF1 in Fig. 4) with either purified vRNP or micrococcus nuclease-treated vRNP (mnRNP) (Fig. 4A). Micrococcus nuclease eliminates vRNA and generates viral proteins free of RNA. His-myc-Tat-SF1 specifically bound to NP among vRNP components when mnRNP was used. This result indicates that Tat-SF1 interacts with NP free of RNA but not NP associated with RNA. Based on this notion, we assumed that Tat-SF1 may have a chaperone-like activity for NP, that is, that Tat-SF1 binds to NP, transfers NP to RNA, and is dissociated from NP during transfer of NP. The addition of increasing amounts of RNA dissociated the Tat-SF1-NP complex (Fig. 4B, lanes 6 and 7). This suggests that NP or the NP-RNA complex was released from Tat-SF1 by the addition of RNA, and the RNA-NP-Tat-SF1 trimeric complex was not formed. Then, we examined the effect of Tat-SF1 on the efficiency of vRNP-NP complex formation (Fig. 4C). A 32P-labeled RNA probe was incubated with recombinant NP in the presence or absence of recombinant Tat-SF1 and was subjected to separation through a 15–35% linear glycerol density gradient. After centrifugation, aliquots were fractionated, and RNA-NP complexes were analyzed by native PAGE. The mobility of RNA-NP complexes was slower than that of the RNA probe alone. We found that formation of vRNA-NP complexes is increased in the presence of Tat-SF1 (Fig. 4C, compare lanes 3–5 with lanes 12–14). The shift of the RNA probe was not found only in the presence of Tat-SF1 (Fig. 4C, lanes 19–27). These results suggest that Tat-SF1 facilitates the formation of vRNA-NP complexes possibly by functioning as a chaperone for NP. This observation is exactly the same as for RAF-2p48 (14), which facilitates the binding of NP to naked viral RNA to form vRNA-NP complexes as active templates for viral RNA synthesis. Tat-SF1 and RAF-2p48 may have a redundant function as NP-interacting factors for the stimulation of viral RNA synthesis (see Discussion).
Fig. 4.
Stimulatory activity of Tat-SF1 in vitro. (A) Interaction between Tat-SF1 and viral proteins. Immunoprecipitation assays were carried out by anti-myc antibody-conjugated agarose beads using purified His-myc-Tat-SF1 (lanes 6–8) and either vRNP (lanes 4 and 7) or mnRNP (lanes 5 and 8). Affinity beads were washed with immunoprecipitation buffer containing 300 mM KCl. Western blot analyses were carried out with anti-NP, -PB1, -PB2, -PA, or -myc antibodies. Input (20%) is shown in lanes 1 and 2. (B) Dissociation of NP-Tat-SF1 complexes by the addition of RNA. An NP-Tat-SF1 complex was reconstituted by mixing purified His-myc-Tat-SF1 (lanes 3 and 5–7) and mnRNP (lanes 4–7). The mixtures were further incubated in the presence of v53-mer RNA [20 ng (lane 6) and 200 ng (lane 7)]. After immunoprecipitation assays using anti-myc antibody-conjugated agarose, Western blot analyses were carried with anti-NP or -myc antibodies. Input (20%) is shown in lane 1. (C) Tat-SF1-mediated NP-RNA complex formation. 32P-labeled RNA probe (v53 mer RNA) mixed with recombinant NP was incubated in the presence of BSA (lanes 1–9) or recombinant Tat-SF1 (lanes 10–18), and v53 mer RNA probe mixed with recombinant Tat-SF1 was incubated in the absence of recombinant NP (lanes 19–27). After sedimentation through a 15–35% glycerol density gradient, fractions were collected from the top of the tube. An aliquot of each fraction was analyzed by PAGE on a 6% polyacrylamide gel, and the RNA was visualized by autoradiography. The unbound RNA probe and NP-RNA complexes are indicated by arrowheads.
Discussion
Here we describe a system for the replication and transcription of an influenza virus genome of negative polarity in yeast cells. With this yeast system, we identify Tat-SF1 as a host factor candidate, which is known as a transcription elongation factor of DNA transcription (30–32). We found that Tat-SF1 stimulates viral RNA synthesis in a cell-free system using vRNP and ApG dinucleotide primers (data not shown). Several splicing proteins may be found to be candidates as host factors for the influenza virus replication process (SI Fig. 6). Some splicing factors are associated with the Pol II (RNA polymerase II)-dependent transcription complex. Recently, it has been reported that influenza virus gene transcription depends on transcriptionally functional Pol II (34). We assume that vRNP could be associated with Pol II complexes, the factors of which may be used as host factors for efficient viral RNA synthesis.
Tat-SF1 may act as chaperone for NP in formation of RNA-NP complexes. In general, nucleic acid-binding proteins containing a basic domain(s) (such as histones) and viral basic proteins (such as histones and NP) tend to aggregate and become inactive in the absence of nucleic acids or appropriate binding proteins (such as molecular chaperones), including histone chaperones (35–37). Because NP is produced as a form free of RNA in infected cells, the interaction of NP with Tat-SF1 may repress the nonspecific aggregation of NP and facilitate the formation of the vRNA-NP complex. Previously, RAF-2p48 was identified as a host-derived chaperone for NP that interacts with NP and stimulates influenza virus RNA synthesis (14). It is possible that Tat-SF1 may have a role similar to that of RAF-2p48 for the stimulation of viral RNA synthesis. NP may use redundant host factors in efficient viral RNA synthesis.
It has been shown that yeast cells support the replication of some positive-strand viral RNA genomes (38). In negative-strand RNA viruses, the expression of viral proteins of vesicular stomatitis virus was observed to depend on primary transcription and translation in yeast cells (39). Here, we show the usefulness of the viral replication system for a negative-strand RNA virus in yeast for the identification and characterization of factors involved in virus replication processes.
To further use the system for screening host factors and an inhibitory drug for the influenza virus, vRNA containing a reporter gene could be convenient. To this end, we tried to replace all viral components consisting of vRNP complexes with viral proteins supplied from exogenously added plasmids (data not shown). We constructed plasmids for the expression of PB1, PB2, PA, and NP under the control of the GAL1 promoter and prepared a model viral RNA (NS-yEGFP RNA) using an in vitro T7 RNA polymerase-directed transcription system (8, 9), in which the yeast-enhanced GFP gene (yEGFP) coding region is sandwiched with the 5′- and 3′-terminal sequences of segment 8 vRNA encoding NS. The expression of yEGFP was detected from NS-yEGFP RNA in yeast cells expressing three viral RNA polymerase subunits and NP (data not shown). However, the expression level of yEGFP was quite low. This system is under improvement in our laboratory.
Materials and Methods
Yeast Strains and Introduction of DNA and vRNP into Yeast Cells.
Yeast strain YPH499 (MATa, ura3–52, lys2–801, ade2–101, trp1–63, his3–200, and leu2–1) was used in all experiments. The sub2 deletion strain was generated by replacement of the entire sub2 ORF with TRP1 (sub2Δ::TRP1 fragment). The lithium acetate–polyethylene glycol method was used for transformation of yeast cells. Introduction of vRNP was performed according to the procedure for transformation with RNA minor modifications (SI Text).
Virus Infection.
Preparation of allantoic fluid from influenza A PR/8 virus-infected embryonated chicken eggs and the infection process has been described (19). HeLa cells were infected at a multiplicity of infection (moi) of 5. Total RNA was prepared by using guanidine methods for RT-PCR (SI Text).
Acknowledgments
We thank Y. Kikuchi and K. Turan for useful discussions. We thank A. Kikuchi and T. Akashi (Nagoya University, Nagoya, Japan) for generous gifts of pRGO1, pRS-513, pRS-317, pRS-315, pYES2, and yEGFP cDNA; R. C. Condit (University of Florida, Gainesville) for pET32aJ3R and pJ3R; and Y. Kawaoka (University of Tokyo, Tokyo) for pcDNA-PB1, pcDNA-PB2, pcDNA-PA, and pHH21.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705856104/DC1.
References
- 1.Bullock PA. Crit Rev Biochem Mol Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 2.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 3.Prentice E, McAuliffe J, Lu X, Subbarao K, Denison MR. J Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholle F, Girard YA, Zhao Q, Higgs S, Mason PW. J Virol. 2004;78:11605–11614. doi: 10.1128/JVI.78.21.11605-11614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. J Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Portela A, Digard P. J Gen Virol. 2002;83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 7.Rochovansky OM, Hirst GK. Virology. 1976;73:339–349. doi: 10.1016/0042-6822(76)90395-0. [DOI] [PubMed] [Google Scholar]
- 8.Luytjes W, Krystal M, Enami M, Pavin JD, Palese P. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka K, Ogasawara N, Yoshikawa H, Ishihama A, Nagata K. Proc Natl Acad Sci USA. 1991;88:5369–5373. doi: 10.1073/pnas.88.12.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 12.Kawaguchi A, Naito T, Nagata K. J Virol. 2005;79:732–744. doi: 10.1128/JVI.79.2.732-744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer D, Molawi K, Martinez-Sobrido L, Ghanem A, Thomas S, Baginsky S, Grossmann J, Garcia-Sastre A, Schwemmle M. J Proteom Res. 2007;6:672–682. doi: 10.1021/pr060432u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Momose F, Basler CF, O'Neill RE, Iwamatsu A, Palese P, Nagata K. J Virol. 2001;75:1899–1908. doi: 10.1128/JVI.75.4.1899-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amorim MJ, Digard P. Vaccine. 2006;24:6651–6655. doi: 10.1016/j.vaccine.2006.05.066. [DOI] [PubMed] [Google Scholar]
- 16.Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, Nagata K. J Biol Chem. 2002;277:45306–45314. doi: 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- 17.Deng T, Engelhardt OG, Thomas B, Akoulitchev AV, Brownlee GG, Fodor E. J Virol. 2006;80:11911–11919. doi: 10.1128/JVI.01565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelhardt OG, Fodor E. Rev Med Virol. 2006;16:329–345. doi: 10.1002/rmv.512. [DOI] [PubMed] [Google Scholar]
- 19.Naito T, Momose F, Kawaguchi A, Nagata K. J Virol. 2007;81:1339–1349. doi: 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panavas T, Serviene E, Brasher J, Nagy PD. Proc Natl Acad Sci USA. 2005;102:7326–7331. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, Ahlquist P. Proc Natl Acad Sci USA. 2003;100:15764–15769. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vreede FT, Jung TE, Brownlee GG. J Virol. 2004;78:9568–9572. doi: 10.1128/JVI.78.17.9568-9572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enami M, Enami K. J Virol. 2000;74:5556–5561. doi: 10.1128/jvi.74.12.5556-5561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noble SM, Guthrie C. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Libri D, Graziani N, Saguez C, Boulay J. Genes Dev. 2001;15:36–41. doi: 10.1101/gad.852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 27.Kikuno R, Nagase T, Suyama M, Waki M, Hirosawa M, Ohara O. Nucleic Acids Res. 2000;28:331–332. doi: 10.1093/nar/28.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Will CL, Luhrmann R. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 29.Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr Mol Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Q, Sharp PA. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- 31.Li XY, Green MR. Genes Dev. 1998;12:2992–2996. doi: 10.1101/gad.12.19.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parada CA, Roeder RG. EMBO J. 1999;18:3688–3701. doi: 10.1093/emboj/18.13.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fong YW, Zhou Q. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 34.Chan AY, Vreede FT, Smith M, Engelhardt OG, Fodor E. Virology. 2006;351:210–217. doi: 10.1016/j.virol.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 35.Okuwaki M, Kato K, Shimahara H, Tate S, Nagata K. Mol Cell Biol. 2005;25:10639–10651. doi: 10.1128/MCB.25.23.10639-10651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okuwaki M, Iwamatsu A, Tsujimoto M, Nagata K. J Mol Biol. 2001;311:41–55. doi: 10.1006/jmbi.2001.4812. [DOI] [PubMed] [Google Scholar]
- 37.Haruki H, Gyurcsik B, Okuwaki M, Nagata K. FEBS Lett. 2003;555:521–527. doi: 10.1016/s0014-5793(03)01336-x. [DOI] [PubMed] [Google Scholar]
- 38.Alves-Rodrigues I, Galao RP, Meyerhans A, Diez J. Virus Res. 2006;120:49–56. doi: 10.1016/j.virusres.2005.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makarow M, Nevalainen LT, Kaariainen L. Proc Natl Acad Sci USA. 1986;83:8117–8121. doi: 10.1073/pnas.83.21.8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yaffe MP, Schatz G. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]