Abstract
Flavin-binding LOV domains are blue-light photosensory modules that are conserved in a number of developmental and circadian regulatory proteins in plants, algae, and fungi. LOV domains are also present in bacterial genomes, and are commonly located at the amino termini of sensor histidine kinases. Genes predicted to encode LOV-histidine kinases are conserved across a broad range of bacterial taxa, from aquatic oligotrophs to plant and mammalian pathogens. However, the function of these putative prokaryotic photoreceptors remains largely undefined. The differentiating bacterium, Caulobacter crescentus, contains an operon encoding a two-component signaling system consisting of a LOV-histidine kinase, LovK, and a single-domain response regulator, LovR. LovK binds a flavin cofactor, undergoes a reversible photocycle, and displays increased ATPase and autophosphorylation activity in response to visible light. Deletion of the response regulator gene, lovR, results in severe attenuation of cell attachment to a glass surface under laminar flow, whereas coordinate, low-level overexpression of lovK and lovR results in a light-independent increase in cell–cell attachment, a response that requires both the conserved histidine phosphorylation site in LovK and aspartate phosphorylation site in LovR. Growing C. crescentus in the presence of blue light dramatically enhances cell–cell attachment in the lovK–lovR overexpression background. A conserved cysteine residue in the LOV domain of LovK, which forms a covalent adduct with the flavin cofactor upon absorption of visible light, is necessary for the light-dependent regulation of LovK enzyme activity and is required for the light-dependent enhancement of intercellular attachment.
Keywords: Caulobacter, LOV domain, photoreceptor, signal transduction, histidine kinase
Proteins that serve as detectors of environmental signals are often modular, containing conserved sensory domains that control diverse signaling outputs (1, 2). One such sensory module is the PAS (Per-ARNT-Sim) domain, which is conserved across all kingdoms of life and is capable of specifically binding a wide range of ligands including heme, flavins, p-coumaric acid, citrate, and other small molecules (3). A subclass of PAS domains, known as LOV domains for their role as sensors of light, oxygen, or voltage, commonly bind a flavin cofactor and function to regulate a number of blue light-dependent processes in plants and fungi (4). These photosensory LOV domains signal by means of a unique photocycle in which photon absorption drives the reversible formation of a covalent adduct between the 4a carbon of the flavin isoalloxazine ring and a conserved cysteine residue (5, 6). Adduct formation is followed by a large structural change at the C terminus of the LOV domain that leads to cell signaling (7, 8). Beyond plants and fungi, dozens of proteins containing LOV photosensory domains have been identified in bacterial species (4, 9). Examples of bacterial LOV photosensors include LOV-phosphodiesterases, LOV-HTH transcription factors, LOV-histidine kinases, and LOV-STAS sigma factor regulators, an example of which has been shown to function as a part of the σB stress response pathway in Bacillus subtilis (10, 11). Among the bacterial LOV photosensors, LOV-histidine kinases are by far the most abundant class (12). The biochemical and biophysical properties and functional role(s) of these photosensory histidine kinases remain largely uncharacterized. However, recent results demonstrating that visible light increases virulence in the mammalian pathogen, Brucella abortus, via a photoactive LOV histidine kinase (13) speak to an important cellular role for this class of two-component signaling proteins.
The two-component family of signal transduction proteins function as the primary signaling systems in bacterial cells (14) and are comprised of (i) sensor histidine kinases and (ii) response regulators. Canonical two-component signaling entails signal-induced autophosphorylation of a histidine protein kinase on a conserved histidine followed by phosphoryl transfer to a conserved aspartate residue on a cognate response regulator protein. Upon modification of their phosphorylation state by a histidine kinase, response regulators go on to modulate cellular physiology or development by either acting directly as transcription factors or through interaction with downstream signaling partners (15).
The observation that LOV histidine kinases are distributed across several bacterial taxa, including many species with no known photobiology (4, 12, 16), suggests that the visible light environment has an unexplored regulatory role in the biology of bacterial cells. One species that is neither photosynthetic, phototactic, nor pigmented, yet encodes a LOV histidine kinase, is Caulobacter crescentus. Here we characterize the biochemical properties and cellular function of a LOV-family photosensory two-component system in C. crescentus consisting of the LOV histidine kinase, LovK, and the single-domain response regulator, LovR. We demonstrate that the purified histidine kinase, LovK, binds flavin, undergoes a reversible photocycle and that its enzyme activity is regulated by visible light. Moreover, we show that the LovK/LovR system and the light environment can act to regulate bacterial cell–surface and cell–cell attachment.
Results
lovK and lovR Are Adjacent Two-Component Genes with Temporally-Correlated Transcription.
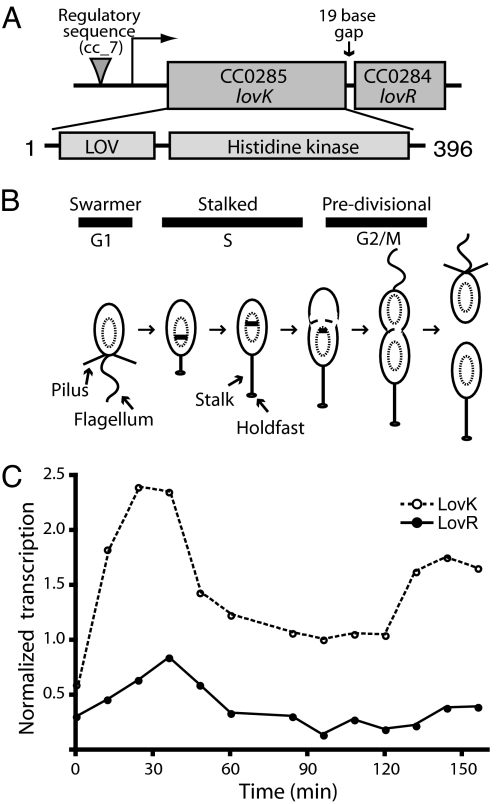
C. crescentus lovK encodes an HWE family (17) histidine protein kinase that is located 17 bp upstream of the gene, lovR, encoding a single-domain response regulator (Fig. 1A). Temporal transcriptional profiling of C. crescentus gene expression across the cell cycle (Fig. 1B) has demonstrated that transcription of lovK and lovR is cell-cycle regulated (18); both genes exhibit correlated expression (Pearson R = 0.92), with maximal expression at the swarmer-to-stalked cell transition and minimal expression during cytokinesis (Fig. 1C). Based on experimental and bioinformatic data, we reannotated the translation start site of lovK [supporting information (SI) Fig. 5] and subsequently identified a conserved GGAACC-N15-CGTT motif 55 bp upstream of the lovK start codon (Fig. 1A). This motif matches a regulatory motif known as cc7, which was previously identified by McGrath and colleagues (18) and demonstrated to have a functional role in transcription. The cc7 motif is upstream of at least 25 other C. crescentus genes that exhibit a qualitatively identical temporal transcriptional profile across the cell cycle.
Fig. 1.
lovK and lovR are organized in an operon whose transcription is temporally regulated across the cell cycle. (A) The gene structure of the lovKR operon (top line) with the cc7 regulatory motif GGAACC-N15-CGTT (18) upstream of lovK indicated with a triangle. The domain structure of the LovK protein is schematized below. (B) Asymmetric division and developmental progression of C. crescentus. The swarmer cell is characterized by a single polar flagellum and polar pili, which are lost during the swarmer-to-stalked transition and replaced by a membranous stalk; at the tip of the stalk is an adhesive holdfast. The G1, S, and G2 phases of the cell cycle are indicated. The chromosome is represented by an oval inside the cell; chromosome replication is indicated by the theta structure. (C) Transcription of lovK (dotted line) and lovR (solid line) are correlated (Pearson R = 0.92) across the cell cycle. Levels of both mRNAs peak during the swarmer-to-stalked transition, fall during DNA replication in the S phase, and rise again during cytokinesis. The time scale is shared for B and C.
The periodic transcription of the lovK/lovR operon, peaking at the swarmer-to-stalked cell transition, provides evidence that the function of the LovK/LovR two-component system is under the larger regulatory control of the C. crescentus cell cycle signaling network. C. crescentus cell attachment requires proper development of cell surface structures, including the holdfast, pili, and flagellum (19). The maximum transcription of lovK and lovR overlaps with the period of polar holdfast biogenesis (20); LovK/LovR may be part of this developmental pathway.
LovK Exhibits a Reversible Photocycle with Slow in Vitro Recovery Kinetics.
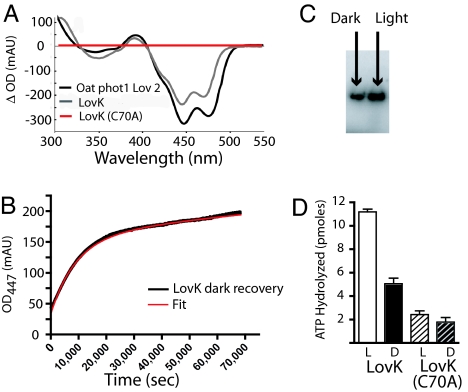
Purified LovK displays a visible absorption spectrum characteristic of oxidized flavoproteins with maximum absorbance at 446 nm and vibronic peaks at 422 nm and 472 nm (Fig. 2A and SI Fig. 6) (9). LovK exhibits a reversible photocycle characteristic of other plant and fungal LOV domains: upon illumination, peaks at 422, 446, and 472 nm bleach and a single band appears at 390 nm (Fig. 2A). Visible absorption spectroscopy (21) and NMR (6) and crystallographic (5) analyses of plant LOV domains have demonstrated that the spectral changes associated with generation of the photoactivated signaling state correspond to the formation of a transient covalent bond between a conserved cysteine residue and the 4a carbon of the flavin isoalloxazine ring. This cysteinyl–flavin adduct ruptures in the dark as the protein recovers back to the ground state. When the conserved cysteine that is predicted to form the adduct in LovK is mutated to alanine (C70A), LovK binds flavin and exhibits an identical dark state spectrum as wild type LovK, but does not undergo any spectral changes upon illumination (Fig. 2A and SI Fig. 6).
Fig. 2.
LovK exhibits a canonical light-dependent LOV absorption spectrum and light regulated kinase activity. (A) Light-minus-dark difference absorption spectrum of LovK is qualitatively identical to that of the oat phototropin-1 LOV2 domain. Light-dependent spectral changes are abolished in the LovK (C70A) mutant. (B) LovK photorecovery in vitro, monitored by the reappearance of the 447-nm peak as the flavin–cysteine adduct ruptures. LovK dark decay is approximated by fitting to the sum of two exponential decay events, indicating that multiple rate constants govern photorecovery. (C) Autophosphorylation of the LovK histidine kinase, detected by autoradiography of an SDS/PAGE gel, is up-regulated by white light. (D) ATPase activity of LovK is up-regulated by white light as measured by monitoring the hydrolysis of [γ-32P]ATP via thin layer chromatography.
Previous studies on LOV domains have demonstrated in vitro dark recovery times ranging from minutes to hours, whereas select LOV proteins, including a LOV histidine kinase of Brucella melitensis (13) and the FKF1 regulator of circadian rhythms in Arabidopsis thaliana (22), do not exhibit any measurable dark recovery after photoexcitation. Purified C. crescentus LovK exhibits slow recovery kinetics from the photoactivated state. A single exponential is not sufficient to fit the recovery data; the data were best fitted as the sum of two exponential decays with recovery half-times of 1.7 h and ≈32 h (Fig. 2B). Given the time limits of these data, the half-life of the second phase is approximate. There may be slower LovK recovery phases, but purified samples were unstable at room temperature and began to precipitate after 12–15 h, which prevented us from accessing these data.
Notably, these kinetic data were collected in the presence of 10 mM imidazole, which dramatically stabilized purified LovK preparations, but accelerates the dark recovery rate of isolated LOV domains (23). Measurement of long time-scale recovery kinetics on purified LovK without imidazole failed due to instability and precipitation of the protein. However, we were able to determine that recovery kinetics under such conditions were qualitatively slower, by at least a factor of 20 (data not shown). If recovery in vivo is unassisted by other factors, these kinetic data suggest that low levels of environmental light are sufficient to generate a long-lived population of photoactivated LovK. However, it should be noted that in the context of the cytoplasm of a bacterial cell, there are high concentrations of imidazole-like molecules (and perhaps other small molecular catalysts) that likely serve to speed the recovery rate from the photoactivated state. Thus the in vitro recovery times reported here may not be relevant to the half-life of the photoactivated signaling state in vivo.
Light Regulates LovK Kinase Activity.
We quantified LovK kinase activity in the light and the dark in two ways: (i) measuring phosphorylated protein separated on SDS/PAGE and (ii) measuring ATP hydrolysis activity of purified LovK via TLC. Purified LovK exposed to ≈200 μE m−2 s−1 of white light for 2 min exhibits 1.7-fold higher autophosphorylation activity than a mock irradiated “dark” sample (Fig. 2C). However, mock irradiated LovK still exhibits significant basal autokinase activity after 12 h of dark recovery. Measuring ATP hydrolysis of purified LovK via TLC supports the SDS/PAGE data. Illumination with ≈200 μE m−2 s−1 of white light increases LovK ATPase activity 2.0-fold relative to mock-irradiated dark samples (Fig. 2D). In the photochemically deficient LovK(C70A) mutant, there is no difference in light-versus-dark ATPase activity. There is some basal ATP hydrolysis activity by LovK (C70A), but it is lower than the “dark” wild-type protein. The elevated activity of dark wild-type protein may be explained by the slow recovery kinetics of LovK, resulting in incomplete photorecovery of the dark wild-type protein after 12 h at 4°C.
LovR Regulates Cell-Surface Attachment.
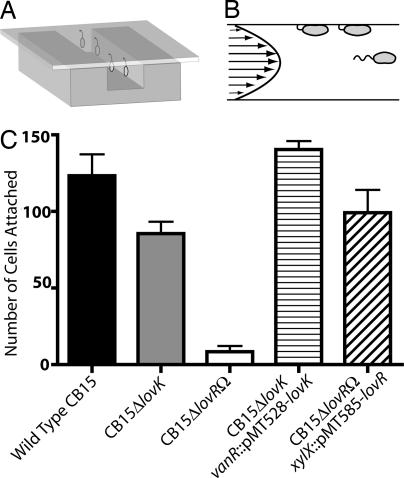
C. crescentus readily attaches to biotic and abiotic surfaces and to other cells in its natural habitat (24). The cellular basis of attachment in C. crescentus is a complex process mediated by surface structures, including the holdfast, pili, and flagellum (19, 25) (see Fig. 1B for an illustration of these structures). Using microfluidic channels (Fig. 3A), we measured cell attachment to a glass surface under laminar flow (Fig. 3B) for the wild-type strain CB15, strains carrying chromosomal deletions of the histidine kinase LovK (CB15ΔlovK), the response regulator LovR (CB15ΔlovRΩ), and strains in which these gene deletions were complemented via expression from inducible promoters (Fig. 3C). Deletion of lovK results in a decrease in the mean number of cells that remain attached to the glass surface under flow relative to wild-type C. crescentus. However, the data are not normally distributed and this decrease is not significant as determined by using a stringent nonparametric ANOVA test (Kruskal–Wallis test, Dunn's post P > 0.05). Deletion of the single-domain response regulator, lovR, results in a severe loss of surface attachment that is highly significant relative to wild-type CB15 (Kruskal–Wallis test, Dunn's post P < 0.001). Xylose-induced expression of lovR from the xylX locus complements the loss of cell attachment in CB15ΔlovRΩ (Fig. 3C).
Fig. 3.
Surface attachment is severely attenuated in a lovR deletion background. (A) Schematic showing attachment of C. crescentus cells inside a microfluidic channel. Cells attach to the glass coverslip via the stalk holdfast. When a flow is applied to the system, cells experience flow forces that are sufficient to lay them flat against the coverslip, permitting individual cell attachment to be quantified via phase contrast microscopy. (B) The flow of a water-based medium in a microfluidic channel of these dimensions is laminar; thus flow velocity approaches zero at the sides of the channel. The arrows inside the parabolic flow profile are a cartoon representation of the decreasing medium flow velocity from the center of the channel to the sides in a two-dimensional cross-section. (C) Bar graph of the number of cells that attached to the glass surface of the channel coverslip and remained attached after 12 ml/min medium flow was applied to the system. Tested strains are labeled on the horizontal axis and the average number of cells attached in a field of view is on the vertical axis with error bars showing 1 standard error of the mean.
Coordinate Overexpression of LovK and LovR Results in Robust Intercellular Attachment.
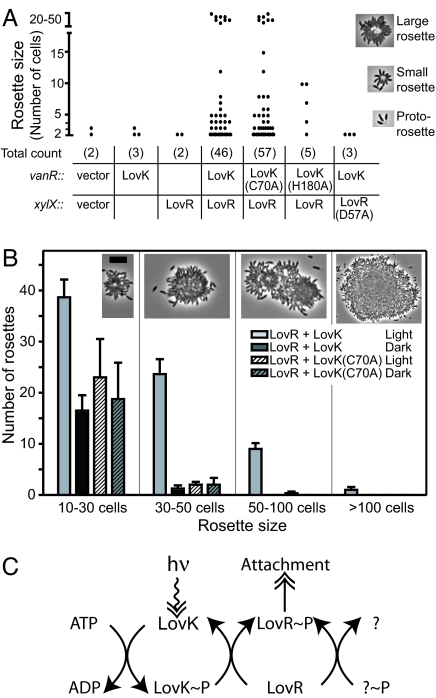
Wild-type C. crescentus attach to each other via their holdfasts to form multicellular structures known as rosettes (see Fig. 4) (24). In the dark, simultaneous low-level overexpression of lovK from a vanillate-inducible promoter and lovR from a xylose-inducible promoter dramatically increases both the number and size of rosettes (Fig. 4A). Strains that overexpress either lovK or lovR individually exhibit no difference in rosette number or size compared with a wild-type empty vector control strain (Fig. 4A). This result demonstrates that individual overexpression of lovK or lovR is not sufficient to elicit an increase in rosette size or number; overexpression of the complete LovK/LovR two-component system is necessary to produce the superattachment phenotype. This phenotype is not dependent on light, as cells were grown completely in the dark.
Fig. 4.
Overexpression of LovK and LovR increases cell-cell adhesion in a light modulated manner. (A) Coordinate, low-level overexpression of both LovK and LovR with intact phosphorylation sites leads to elevated levels of cell-cell adhesion in dark-grown cultures. The proteins overexpressed in each strain are indicated below the plot. LovK overexpression variants or an empty plasmid were integrated into the vanR locus and LovR variants or empty plasmid were integrated into the xylX locus. Total numbers of rosettes observed for each strain are indicated in parentheses. Illustrative examples of the three rosette types are shown on the right. (B) Light-dependent enhancement of cell-cell attachment in a LovK/LovR overexpression background requires the C70 residue of LovK. Average number of large rosettes binned by approximate size; error bars indicate standard error of the mean. Representative images of rosettes in each size class are shown above each bin. (Scale bar, 5 μm, except for the largest rosette where it represents 2.5 μm.) (C) A proposed model of the LovK/LovR signaling pathway. Gene deletion and gene overexpression data demonstrate that LovK and LovR are regulators of cell attachment. Deletion of lovR nearly abolishes attachment to a glass surface, whereas deletion of lovK only modestly attenuates attachment, suggesting there may be an additional regulator of LovR (shown as a question mark in this panel). Mutagenesis of the conserved phosphorylation sites in this two-component system provides evidence that LovK and LovR phosphorylation is necessary for the large increase in intercellular attachment during coordinate LovK/LovR overexpression. Light acts to up-regulate both the kinase activity of LovK in vitro, and cell–cell attachment in vivo; both of these responses require cysteinyl-flavin adduct formation in the LOV domain of LovK. Filled arrows indicate phosphotransfer events. Double-headed arrows indicate signal input and cellular output.
“Super Attachment” Requires the Conserved Phosphorylation Sites of LovK and LovR.
To test whether the conserved sites of histidine phosphorylation in lovK and aspartate phosphorylation in lovR are necessary for the superattachment phenotype, we created site-directed overexpression mutants that encoded a histidine to alanine mutation in LovK (H180A) or an aspartate to alanine mutation in LovR (D57A). Simultaneous overexpression of wild-type LovK and LovR (D57A) failed to exhibit any difference in rosette size or number compared with a wild-type empty-vector control. Overexpression of LovK (H180A) and wild-type LovR resulted in a total number of rosettes comparable to that of a wild-type empty-vector control, although there were two mid-sized rosettes recorded (Fig. 4A). Nevertheless, rosestte size and number were still significantly lower than the wild-type LovK/LovR double overexpression strain (Fig. 4A). Thus, both the histidine phosphorylation site of LovK and aspartate phosphorylation site of LovR are necessary to elicit the superattachment phenotype during LovK/LovR overexpression.
LovK Cysteinyl-Flavin Adduct Formation is Required for Light-Dependent Accentuation of Cell Attachment.
To determine whether the conserved cysteine required for cysteinyl-flavin adduct formation (Fig. 2A) was necessary for the superattachment phenotype described above, a site-directed mutant that encoded a cysteine to alanine mutation in LovK (C70A) was constructed. A strain overexpressing LovK (C70A) from the vanillate locus and wild-type LovR from the xylose locus exhibited large and abundant rosettes in the dark comparable to those in the wild-type LovK/LovR overexpression strain (Fig. 4 A and B). This result indicates that unlike LovK-H180 and LovR-D57, LovK-C70 is not required for super attachment during LovK/LovR overexpression, confirming the light independence of this phenotype.
We next sought to examine the role of light in the LovK/LovR signaling pathway. For optimal photoexcitation of LovK in vivo, cells were cultured under blue light (see Materials and Methods). Relative to dark-grown cells, cultures grown in the light contained more total rosettes and displayed extremely large rosettes (Fig. 4B). Moreover, light exposure results in large mats of cells on the surface of cultures overexpressing LovK/LovR (SI Fig. 7). This light-dependent accentuation of cell-cell attachment and cellular mat formation requires the C70 residue of LovK, providing evidence that light-driven adduct formation, and subsequent up-regulation of LovK autophosphorylation controls this response (Fig. 4B and SI Fig. 7).
Discussion
To understand the molecular and cellular basis of environmental signal detection and integration in bacteria, one must first define the physical and chemical signals that are relevant to the biology of a bacterial cell. The work presented herein demonstrates that visible light can act as a regulatory signal that increases the biochemical activity and cellular signaling output of the LovK/LovR two-component system in C. crescentus. Genetic loss-of-function data show that LovR has a basal role in the control of cell attachment in the absence of light. Whereas deletion of LovR nearly abolishes attachment, deletion of LovK only modestly attenuates attachment, suggesting that there may be additional regulators of LovR activity (Fig. 4C). Coordinate overexpression of LovK and LovR results in strongly elevated intercellular attachment in a C. crescentus monoculture; this response requires the conserved phosphorylation sites of both LovK and LovR. Light-dependent accentuation of this cell–cell superattachment response requires a conserved cysteine residue that forms a covalent cysteinyl–flavin adduct in the LovK protein and is necessary for light-regulated LovK enzyme activity.
What, then, is the relevance of blue light to the biology of this species? C. crescentus is an oligotrophic bacterium that thrives in dilute freshwater environments, and is capable of living at various depths in the water column. We propose that light is an environmental signal that can cue the Caulobacter cell on where it is positioned in the water column. Blue wavelengths of visible light are particularly well-suited to act as signals for aquatic bacteria, as light in the blue region of the visible spectrum penetrates the water column to a greater depth than lower energy yellow and red light (26). One possible function of blue light could be related to nutrient availability. Specifically, the total nutrient pool is less abundant at the surface of freshwater lakes than in the benthic sediment (26, 27), and up-regulating the capacity to attach to limited nutrient detritus at the surface (where the cell is exposed to more light) could provide a growth advantage. Homologous two-component systems to LovK/LovR are present in the aquatic oligotrophs Erythrobacter litoralis and Novosphingobium aromaticivorans, suggesting that light-modulated cell attachment may be a general adaptive strategy for bacteria in such ecological niches. Additionally, related LOV histidine kinases are found in other aquatic species such as Magnetospirillum magnetotacticum, Aurantimonas sp., Parvularcula bermudensis, and Oceanicola granulosus.
LOV histidine kinases are also conserved in the genomes of plant and human pathogens such as Pseudomonas syringae and Brucella abortus, respectively (16). Cell attachment is intimately tied to virulence in many bacterial pathogens (28). It will be interesting to test whether light-regulated cell attachment is involved in Brucella virulence (13) and to determine whether light and LOV histidine kinases function generally as regulators of cell attachment or cell surface properties, or control more diverse cellular outputs.
Materials and Methods
Cloning, Overexpression, and Purification of LovK.
The gene encoding the predicted LOV-containing histidine protein kinase, lovK (gene number CC0285) was PCR amplified from C. crescentus genomic DNA (see SI Fig. 5 for gene annotation information). The PCR product was gel purified, digested with EcoRI and HindIII (NEB, Ipswich, MA), and cloned into the overexpression vector pET28a (Novagen, Madison, WI). pET28a-lovK was transformed into Rosetta(DE3)pLysS (Novagen) to create the overexpression strain FC410. FC410 was grown at 37°C in 1 liter of LB broth and 30 μg/ml of Kanamycin to an OD600 of 0.4 (1 cm path length), at which point the temperature was lowered to 20°C and the cells were induced with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h. Cells were lysed in 20 mM Tris·HCl (pH 7.5), 500 mM NaCl, 1 mM EDTA, and 10 mM imidazole. The recombinant His-6–LovK protein was purified by using Ni2+ Chelating Resin (Amersham Pharmacia, Piscataway, NJ). Affinity purified LovK was dialyzed overnight against 20 mM Tris·HCl (pH 7.5), 20 mM NaCl, 10 mM imidazole and further purified via Q-Sepharose (Amersham Pharmacia) anion exchange. A mutant version of pET28a-LovK in which the cysteine residue required for photoactivity was mutated to alanine (LovK-C70A) was generated by using PCR based site-directed mutagenesis (see SI Table 1 for a complete list of strains used in this study).
Visible Absorbtion Spectroscopy on LovK and LovK(C70A).
Absorption spectra of LovK and LovK(C70A) were measured on a Shimadzu UV-1650 spectrophotometer using a quartz cuvette with a 1 cm path length. Purified LovK was kept in the dark until data collection and photoexcited by using a white tungsten lamp (20-s exposure, 2 mW·cm−2). The rate of recovery from the photoactivated state was determined by measuring the dark-state increase at A447 at intervals of 300 s (23°C). After 12 h, the protein began to precipitate, which created background scattering problems that hindered further measurement. Data were fitted using a two-phase exponential decay function (the sum of two exponential decays) in Prism (GraphPad Software, San Diego, CA).
LovK Kinase Activity Assays.
After purification and concentration, LovK was kept in the dark at 4°C for 12 h. TLC ATPase assays were performed with 50 μM LovK dimer (ε = 11.2 mM−1 cm−1) in buffer containing 2 mM Tris·HCl (pH 7.5), 2 mM MgCl2, 1 mM DTT, 19 μM ATP, and 1 μM [γ-32P]ATP. Kinase reactions were assembled under dim red light at room temperature and either run in red light or exposed to 200 μE m−2 s−1 fluorescent white light for the duration of the reaction. After 20 min, reactions were quenched with 0.5 volumes of 12 N formic acid and kept on ice for 10 min. Reactions were spun for 2 min at maximum speed in a benchtop centrifuge and dropped onto polyethyleneimine cellulose TLC plates (EMD Chemicals), blotted with 10 mM ADP, and run for 7 min in 1 N LiCl, 1 N formic acid. Spots corresponding to unhydrolyzed [γ-32P]ATP and free [32P] phosphate were detected by using a STORM 860 Molecular Imager and quantified with ImageQuant 5.2 software (Molecular Dynamics). Reactions were performed in triplicate, and blanked with a mock reaction containing no LovK, performed in the light. ATP hydrolysis in the no protein blank was subtracted from each reaction. The SDS/PAGE gel kinase activity assay was run under the following conditions: 2 mM Tris·HCl (pH 7.5), 2 mM MgCl2, 2 mM DTT, 200 μM ATP, 0.2 μM [γ-32P]ATP. Photorecovered LovK (50 μM) was added to this mix in the dark, to a final volume of 10 μl. One sample was kept in the dark and one was exposed to fluorescent white light (200 μE m−2 s−1) for 2 min. After 2 min, reactions were quenched with 10 μl of quenching buffer (0.4 M Tris·HCL, pH 10.0/6.6% SDS/1 mM 2-mercaptoethanol/20% glycerol) and kept on ice until being loaded onto a 4–20% gradient SDS gel and run at 150 V at 4°C. The gel was rinsed in deionized H2O and imaged as described above.
lovK and lovR Overexpression Strains and Gene Deletions.
lovK was amplified from genomic DNA, cloned into the NdeI and KpnI sites of vector pMT528 (gift from M. Thanbichler, MPI, Marburg, Germany) and transformed and integrated into the vanR locus of Caulobacter CB15 to generate a vanillate-inducible lovK strain (FC437). lovR (CC0284) was amplified from genomic DNA, cloned into the NdeI and EcoRI sites of the vector pMT585 (gift from M. Thanbichler), and transformed and integrated into the xylX locus of Caulobacter CB15 to generate a xylose-inducible lovR strain (FC375). Electrocompenent cells of FC375 were prepared and pMT528-lovK was transformed and integrated into this cell line to create a Caulobacter CB15 lovK/lovR double–inducible overexpression strain (FC438).
A mutant version of pMT528-LovK that substituted an alanine for the conserved histidine phosphorylation site (H180A) and a mutant version of pMT585-lovR that substituted an alanine for the conserved aspartate phosphorylation site (D57A) were generated via PCR-based site-directed mutagenesis. In addition, pET28a-lovK(C70A) (see above) was subcloned into pMT528 to generate a pMT528-lovK(C70A) mutant overexpression vector. All vectors for in vivo overexpression of wild-type and mutant versions of lovK and lovR were transformed into Caulobacter via electroporation.
Gene deletions of lovK (CC0285) and lovR (CC0284) were generated as follows: two PCR products extending ≈500 bp from either side of the gene to be deleted were amplified from genomic DNA using KOD Hot Start Polymerase (Novagen) and cloned into pNPTS138 (see SI Table 1). An Ω cassette encoding spec/strep resistance was inserted into the pNPTS138-lovR knockout vector. For lovK deletion, the pNPTS138- deletion vector was transformed into wild-type Caulobacter strain CB15 and gene replacement was performed by a two-step sacB counterselection procedure. Colony PCR using a set of primers flanking each deleted gene was used to confirm the presence of the deletion. The lovK deletion is in-frame to avoid polar effects on lovR expression and encodes the first 6 (MEDYSE) and the last 7 aa (LPINGTQ) of the LovK protein (strain FC455). lovR was originally knocked out in a CB15N background, which is deficient in holdfast synthesis and attachment; CB15NΔlovR (strain FC15) contains the first 20 nucleotides, an omega cassette conferring spec/strep resistance, and the last 18 nucleotides of lovR. This marked CB15NΔlovR Ω mutant strain was transduced with phage φCr30 into a wild-type CB15 background to generate CB15ΔlovRΩ (FC301). FC455 and FC301 were complemented with integrating plasmids that carried vanillate- or xylose-inducible lovK and lovR, respectively. These plasmids, from strains FC432 and FC364, were transformed into the deletion backgrounds via electroporation.
Microfluidic Microscopy and Quantitation of Glass Surface Attachment.
Microfluidic channels measuring 200 μm wide by 50 μm deep by 2 cm long were made with polydimethylsiloxane (PDMS, Sylgard Brand 184 Silicone Elastomer kit). The PDMS and glass coverslip were cleaned and sealed by using a Plasma Prep II plasma cleaner (SPI Supplies). Sodium hydroxide (2 M solution), ethanol, and water were sequentially flowed into the channels to clean the interior before cell loading. Individual cell colonies of FC19, FC301, FC455, FC589, and FC590 (SI Table 1) were taken from a PYE-agar plate and grown overnight in 5 ml peptone/yeast extract medium (PYE) at 30°C, diluted 1:25 in fresh PYE plus 0.5 mM vanillate and 0.15% xylose, and re-grown to 0.15 OD660. Cells were then loaded into the microfluidic channel and incubated for an additional 30 min at 30°C before imaging. A Harvard Apparatus PHD2000 infuser was used to induce a constant flow of PYE medium at 12 μl/min for the duration of imaging. Cells grown on three separate days were imaged in seven different fields of view per day with a Leica DM5000 at 630× magnification in phase contrast mode. Images were captured with a Hamamatsu Orca-ER digital camera. ImageJ (National Institutes of Health, Bethesda, MD) was used for image processing and cell counting. The data were not normally distributed, and Kruskal–Wallis nonparametric ANOVA with a Dunn's post hoc test was used to detect statistically significant differences in attachment between strains.
Quantification of Cell–Cell Attachment and Cell Growth Under Blue Light.
Cells were initially grown to saturation in M2 media supplemented with 0.15% xylose and 0.5 mM vanillate in the presence of selecting antibiotics (25 μg/ml spectomycin and 5 μg/ml streptomycin for strains carrying pMT528 based constructs, and 5 μg/ml kanamycin for strains carrying pMT585 based constructs) and diluted 100-fold into 10 ml of the same medium without antibiotics. To assess differences between overexpression strains, the diluted cultures were grown to saturation in a roller at 30°C in the dark. Cells were viewed by phase contrast microscopy using the microscope described above. One microliter of culture was spread between a coverslip and a slide, and rosette number and size were counted in 10 randomly selected fields of view (130 μm × 100 μm). Rosettes are groups of cells attached at the holdfast end of the stalk. Thus small groups of two to five cells attached at the tips of the stalk were counted. Cells touching in other ways were not counted. Scatter plots were generated from the distributions of rosette sizes. To assess differences between light and dark grown cultures, cells were treated the same as above except that the second growth was conducted in a 30°C shaker at 230 RPM either wrapped in foil or ≈2 cm from a blue light source. The light source consisted of an array of blue AlGaInP LEDs (20° viewing angle, 8000 mcd, 468 nm λmax at 3.4 V) powered by a 3.3 V, 4 A AC adaptor. The light power at the position in the tube rack where cells were cultured was 1.3 mW·cm−2 (New Focus Power Meter, San Jose, CA). When cultures reached an OD660 of 0.9–1.0, cells in the liquid were gently transferred with a Pasteur pipette to a clean sterile tube. The cells remaining in the tubes loosely associated with the surface of the glass were photographed with a Canon Rebel XTi digital camera. Cells in 1 μl of the transferred media were visualized as described except that rosettes in different size classes were counted in 1 × 5 mm areas of the slide. At least three different areas were counted to ensure representative counting. Each area was treated independently. Graphs were generated by using Prism (GraphPad).
Supplementary Material
Acknowledgments
We thank Lucy Shapiro, Harley McAdams, John Kennis, Winslow Briggs, Trevor Swartz, and Devaki Bhaya for helpful discussion over the course of this study, Patrick McGrath for assistance with promoter motif identification, Brett Engelmann and Cara Boutte for help with data collection, the Scherer and Ismagilov Groups for assistance with microfluidic construction, the Sosnick and Moffat Groups for use of the LED illuminator and spectrophotometers, and Ling Chan for experimental guidance. We also thank the anonymous reviewers, whose criticism and suggestions greatly improved this manuscript. E.P. is funded by National Institutes of Health Training Grant T32 GM007183-32. S.C. is funded by a Beckman Young Investigator Award and a grant from the Mallinckrodt Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705887104/DC1.
References
- 1.Galperin MY. Environ Microbiol. 2004;6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pawson T, Nash P. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 3.Taylor BL, Zhulin IB. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crosson S, Rajagopal S, Moffat K. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 5.Crosson S, Moffat K. Plant Cell. 2002;14:1067–75. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salomon M, Eisenreich W, Durr H, Schleicher E, Knieb E, Massey V, Rudiger W, Muller F, Bacher A, Richter G. Proc Natl Acad Sci USA. 2001;98:12357–12361. doi: 10.1073/pnas.221455298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harper SM, Christie JM, Gardner KH. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 8.Harper SM, Neil LC, Gardner KH. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 9.Losi A. Photochem Photobiol Sci. 2004;3:566–574. doi: 10.1039/b400728j. [DOI] [PubMed] [Google Scholar]
- 10.Avila-Perez M, Hellingwerf KJ, Kort R. J Bacteriol. 2006;188:6411–6414. doi: 10.1128/JB.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaidenko TA, Kim TJ, Weigel AL, Brody MS, Price CW. J Bacteriol. 2006;188:6387–6395. doi: 10.1128/JB.00691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briggs WR. J Biomed Sci. 2007;14:499–504. doi: 10.1007/s11373-007-9162-6. [DOI] [PubMed] [Google Scholar]
- 13.Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, et al. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 14.Robinson VL, Buckler DR, Stock AM. Nat Struct Biol. 2000;7:626–633. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- 15.Stock AM, Robinson VL, Goudreau PN. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 16.Losi A. In: Flavins: Photochemistry and Photobiology. Silva E, Edwards AM, editors. Dorchester, UK: RCS; 2006. pp. 217–269. [Google Scholar]
- 17.Karniol B, Vierstra RD. J Bacteriol. 2004;186:445–453. doi: 10.1128/JB.186.2.445-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, Tan MH, Hillson NJ, Hu P, Shapiro L, McAdams HH. Nat Biotechnol. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- 19.Bodenmiller D, Toh E, Brun YV. J Bacteriol. 2004;186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levi A, Jenal U. J Bacteriol. 2006;188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 22.Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 23.Alexandre MTA, Arents JC, van Grondelle R, Hellingwerf KJ, Kennis JTM. Biochemistry. 2007;46:3129–3137. doi: 10.1021/bi062074e. [DOI] [PubMed] [Google Scholar]
- 24.Poindexter JS. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Entcheva-Dimitrov P, Spormann AM. J Bacteriol. 2004;186:8254–8266. doi: 10.1128/JB.186.24.8254-8266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldman CR, Horne AJ. Limnology. Columbus, OH: McGraw–Hill; 1983. [Google Scholar]
- 27.Lampert W, Sommer U. Limnoecology: The Ecology of Lakes and Streams. Oxford: Oxford Univ Press; 1997. [Google Scholar]
- 28.Donlan RM, Costerton JW. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.