Abstract
The genome of the plant-colonizing bacterium Pseudomonas fluorescens SBW25 harbors a subset of genes that are expressed specifically on plant surfaces. The function of these genes is central to the ecological success of SBW25, but their study poses significant challenges because no phenotype is discernable in vitro. Here, we describe a genetic strategy with general utility that combines suppressor analysis with IVET (SPyVET) and provides a means of identifying regulators of niche-specific genes. Central to this strategy are strains carrying operon fusions between plant environment-induced loci (EIL) and promoterless ′dapB. These strains are prototrophic in the plant environment but auxotrophic on laboratory minimal medium. Regulatory elements were identified by transposon mutagenesis and selection for prototrophs on minimal medium. Approximately 106 mutants were screened for each of 27 strains carrying ′dapB fusions to plant EIL and the insertion point for the transposon determined in approximately 2,000 putative regulator mutants. Regulators were functionally characterized and used to provide insight into EIL phenotypes. For one strain carrying a fusion to the cellulose-encoding wss operon, five different regulators were identified including a diguanylate cyclase, the flagella activator, FleQ, and alginate activator, AmrZ (AlgZ). Further rounds of suppressor analysis, possible by virtue of the SPyVET strategy, revealed an additional two regulators including the activator AlgR, and allowed the regulatory connections to be determined.
Keywords: functional genomics, gene expression, gene regulation, phytosphere, Pseudomonas fluorescens
The function of bacteria in natural environments is poorly understood. If bacteria were the size of birds and possessed similar morphological complexity, then progress could be made by observation alone. In the absence of readily observable phenotypes, microbiologists have taken to detecting changes in patterns of gene expression. Genes expressed in one environment, but not in another, are likely to encode traits relevant to the former environment but not the latter. Understanding the biological significance of these traits, their contribution to ecological performance, and the regulatory networks that control their expression is central to understanding the function of bacteria in the wild.
Pseudomonas fluorescens SBW25 is a rhizosphere colonizing bacterium whose survival at the plant–soil interface depends on ability to integrate a multiplicity of environmental stimuli. Insight into the causes of the bacterium's ecological success have come from application of promoter-trapping strategies, such as IVET (in vivo expression technology) (1), which have led to the identification of genes displaying elevated levels of expression in the plant environment (2–9). Although the biological function and ecological significance of a number of these genes has been determined, significant progress has been hampered because (by definition) the genes of interest express no phenotype in vitro. Progress therefore requires insight into environmentally relevant regulatory circuits and the connections between the components of this circuitry and the traits that determine ecological performance.
Here, we describe a broadly applicable extension of the IVET strategy that allows identification of regulators, both positive and negative, of environment-induced loci (EIL) (10). By manipulating expression of regulators, we show that insight into the biological function of EIL can be obtained along with an understanding of the regulatory networks that determine gene expression in the wild.
Results
SPyVET Screening for Regulators Controlling Expression of Plant Environment-Induced Loci (EIL).
The screen to identify regulators of EIL from P. fluorescens SBW25 [supporting information (SI) Fig. 5] is based on SBW25ΔdapB carrying a chromosomally integrated fusion between an EIL and a promoterless copy of dapB (EIL-′dapB; see Fig. 1) (4, 10, 11). Such a strain is auxotrophic for diaminopimelate (DAP) and lysine in vitro, but prototrophic in the plant environment, where signals activating the EIL (and thus the promoterless copy of dapB) are present. Previous P. fluorescens SBW25 IVET screens have identified a range of EIL, including genes for nutrient acquisition and metal homeostasis, secretion, regulation, stress response, and attachment and surface colonization (2–9). A recent large-scale IVET screen of ≈40,000 fusions has completed the screening process and identified 181 EIL (P.B.R., unpublished data).
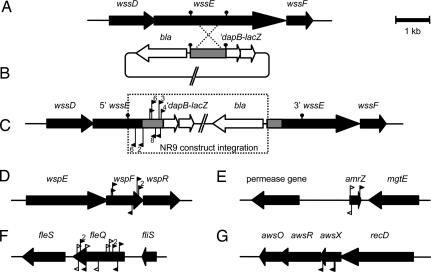
Fig. 1.
Genomic context of an IVET fusion (EIL) and its regulators. (A) Arrangement of wssDEF genes in SBW25 where IVET fusion NR9 (wssE) was located. (B) NR9 (wssE) pIVETD construct. The gray box represents a SBW25 Sau3AI library fragment containing 994 bp of wssE, which was cloned into the BglII site of pIVETD (′dapB, ′lacZ, and bla genes are shown). The position of the Sau3AI sites that generated this fragment are indicated by filled circles. (C) Gene arrangement after homologous recombination between the wssE fragment of the NR9 (wssE) pIVETD construct and its cognate chromosomal copy (cross-over represented by dotted lines). (C–G) The locations of transposons in NR9 (wssE) TAF strains. Transposon insertion sites are represented by triangles pointing from the I-end (IE) of the transposon in the direction of the nptII promoter activity (see also SI Fig. 6). Filled triangles represent IS-Ω-Km/hah insertions, and open triangles represent miniTn5Km insertions. Multiple transposon insertions are enumerated above or below the relevant triangle. Transposon insertions in the following loci resulted in activation of the NR9 wssE::′dapB′lacZ fusion: wss locus (wssDEF PFLU0303-0305 shown) (C), here, promoter activity from the either end of the transposon directly activates ′dapB; wsp locus, transposon insertions disrupt wspF (PFLU1224) but have nonpolar effects and activate wspR (PFLU1225) (D); amrZ (PFLU4744) (E); fleQ (PFLU4443) (F); and awsX (PFLU5211) (G).
For each IVET fusion strain, suppressor mutants were sought by using miniTn5Km (12) and, in a separate experiment, IS-Ω-Km/hah (13) (SI Fig. 6). Transposon-carrying recipients were plated onto minimal M9 medium lacking DAP and lysine, which provided selection for mutants in which the presence of the transposon resulted in conversion from DAP− to DAP+. There are three possible causes for the transcriptional activation of dapB: first, the transposon (of either type) may disrupt (inactivate) a negative regulatory element of the EIL; second, IS-Ω-Km/hah may stimulate transcription of a transcriptional activator of an EIL [by virtue of the internal nptII promoter (SI Fig. 6)]; third, and least interestingly, IS-Ω-Km/hah may directly activate transcription of promoterless dapB by inserting immediately upstream of ′dapB (in the EIL itself). Because the EIL-′dapB fusion is active in each of the three classes of strains, we designated all these strains TAF (transcriptionally active fusion) mutants.
The “suppressor IVET” (SPyVET) strategy is most effectively applied to EIL-′dapB strains that show no growth on minimal M9 medium. Thus, from an initial set of 181 EIL-′dapB fusion strains, 51 showed a complete absence of growth on minimal M9 medium, and 27 of these exhibited no background growth when plated at high cell density (≈105 per cm2). These 27 fusion strains (SI Table 2) were further checked to ascertain the rate of spontaneous mutation to prototrophy (spontaneous TAFs). In every case, it was ≈102 to 103 times less frequent than transposon-generated mutants (SI Table 2). No spontaneous SBW25ΔdapB prototrophs were observed. This demonstrates that an EIL-′dapB fusion is an essential prerequisite for the TAF phenotype.
The genome of SBW25 was comprehensively screened for regulators of EIL by mutagenesis: for each fusion strain, 105 to 106 mutants were screened for prototrophy (yielding between 8- and 750-fold coverage of the 6,007 predicted ORFs in the genome). This led to the identification of between 4 and 363 prototrophic mutants (SI Table 2) for each fusion strain. Arbitrary Prime-PCR and sequencing of between 4 and 135 randomly selected TAF mutants per fusion strain led to identification of the transposon insertion point and orientation in each of 2,292 TAF strains (Fig. 2 and SI Table 3; precise insertion points are supplied in Artemis format (SI Dataset 1) for visualization on the complete SBW25 genome sequence).
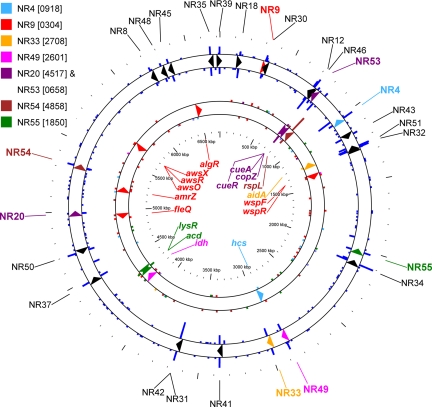
Fig. 2.
Location of plant EIL and related regulatory genes in the SBW25 genome. Two concentric rings are used to show the position on the circular SBW25 genome (6,722,539 bp) of the plant EIL (triangles in outer ring) and the regulators (triangles in inner ring) described in this study. On the outer ring, colored triangles represent plant EIL for which regulators were characterized (see the color key for associated PFLU ORF designations), and all transposon insertions screened during this study are represented by blue lines around the outer circle. The length of each blue line is proportional to the number of insertions per 100-bp window (highest density was 42 insertions per 100 bp). Transposons orientated so that the promoter nptII faces clockwise are shown on the outer circle and counterclockwise on the inner circle. On the inner ring, colored triangles represent the regulators reported in this study (as labeled), and the colored lines indicate regulator-specific transposon insertions (i.e., excluding those adjacent to a dapB fusion) reported as density per 100-bp window.
Most transposon insertions resulting in TAF mutants were located near the EIL-′dapB integration region (Fig. 1C; discussed below), but discrete clusters of insertions were located in loci elsewhere on the genome. Loci containing multiple independent insertions were further characterized after confirming the transposon location by PCR. Loci that received only a single insertion were generally not studied further because of the possibility that their phenotype resulted from a second-site spontaneous mutation. Of the 27 EIL fusions, candidate regulators were identified for eight strains (Fig. 2 and Table 1). By way of example, the genomic location of putative regulators for EIL-′dapB fusion strain NR9 (which contains a promoterless dapB fusion to wssE) is illustrated in Fig. 1.
Table 1.
Effect of regulators on the transcriptional activity of plant EIL in P. fluorescens SBW25
| TAF | EIL fusion† (PFLU no.) | Regulator‡ (PFLU no.) | LacZ activity relative to control§ |
Regulation | |
|---|---|---|---|---|---|
| EIL + regulator | TAF + regulator | ||||
| NR4.P03 | NR4 membrane protein (0918) | hcs (2629) | 1.15 ± 0.37* | 0.99 ± 0.27* | A |
| NR9.P5 | NR9 (wssE) cellulose | fleQ (4443) | 0.03 ± 0.13 NS | −1.42 ± 0.27* | R |
| NR9.P6 | synthase (0304) | awsR (5210) | 1.08 ± 0.16** | −0.07 ± 0.33 NS | A |
| NR9.P6 | awsX (5211) | 0.01 ± 0.29 NS | −2.29 ± 0.18** | R | |
| NR9.7 | wspF (1224) | −0.16 ± 0.14 NS | −1.67 ± 0.19** | R | |
| NR9.20 | amrZ (algZ) (4744) | −0.24 ± 0.11 NS | −2.49 ± 0.31** | R | |
| NR20.4 | NR20 permease (4517) | cueA (0658) | 0.36 ± 0.39 NS | 1.23 ± 0.52 NS | — |
| NR20.4 | cueR (0657) | 1.17 ± 0.23* | 0.82 ± 0.81 NS | A | |
| NR20.4 | copZ (0660) | 0.75 ± 0.13** | 0.87 ± 0.77 NS | A | |
| NR33.95 | NR33 (pinA) nitrilase (2708) | aidA (1102) | 3.65 ± 0.41** | −0.20 ± 0.12 NS | A |
| NR49.10 | NR49 membrane protein (2601) | idh (3809) | −5.02 ± 2.54 NS | 0.07 ± 0.10 NS | R¶ |
| NR53.10 | NR53 (cueA) copper | cueA (0658) | 2.74 ± 0.39** | −0.34 ± 0.17 NS | A |
| NR53.10 | transporter (0658) | cueR (0657) | 2.59 ± 0.47** | −1.75 ± 0.24** | A/R |
| NR53.10 | copZ (0660) | −0.35 ± 0.77 NS | −1.57 ± 0.14** | R | |
| NR54.43 | NR54 response regulator (4858) | rspL (0709) | 3.85 ± 0.30** | 0.60 ± 0.31 NS | A |
| NR55.30 | NR55 response regulator (1850) | acd (3884) | −2.61 ± 0.40** | −2.47 ± 0.51* | R |
| lysR (3885) | 1.45 ± 0.11** | 1.22 ± 0.40* | A | ||
P values after one-way ANOVA analysis are: *, P < 0.05; **, P < 0.01; NS, no significant difference. LacZ activity was normalized against control data, where all four replicates of the control strain (WT or mutant containing a pBroadgate-D control plasmid) were converted to an activity of one. All test values, either WT strain or transcriptionally activated fusion (TAF) strain carrying regulator genes, were divided by the corresponding control value (from the same 96-well plate), and all values were transformed to log2 values. The control values (always one before transformation) equal zero, and test values below zero indicate lower activity compared with the control strain, whereas values above zero indicate higher activity. Strains NR4, NR33, and NR54 are examples of a mutant where the transposon is located upstream of the listed regulator and activates the positive regulator. A, activator; R, repressor.
†NR EIL fusion number, the predicted gene product and the SBW25 genome sequence gene identifier in parentheses.
‡The gene expressed in each EIL and TAF strain (SBW25 gene identifier number in parentheses).
§LacZ values are the mean of four replicates and standard error of mean.
¶Complementation of the idh NR49.10 mutant was not achieved, but the interruption of idh by five independent transposon insertions in both orientations and the observation that mutant NR49.10 exhibits a 6-fold higher lacZ activity than the WT strain (data not shown) strongly argues in favour of idh encoding a repressor.
Prediction and Functional Confirmation of Candidate Regulators.
The majority of transposon insertions mapped near the dapB integration point confirming nptII promoter activity from the I-end (IE) of IS-Ω-Km/hah (13). Promoter activity was also evident from the O-end (OE; SI Fig. 6). RACE (rapid amplification of cDNA ends) analysis indicated that transcription started ≈187 bases in from the IE and 4 bases in from the OE of IS-Ω-Km/hah (data not shown). These insertions were not further considered.
Unique regulatory candidate loci identified for each of the eight fusion strains are listed in Table 1. To experimentally confirm the regulatory roles of candidate genes and rule out the possibility of polar effects, the influence of each candidate regulator gene on expression of the EIL-′dapB fusion in both the original fusion strain [e.g., NR9 (wssE)] and its respective TAF mutant (e.g., NR9.5) was tested. Each gene was cloned downstream of a constitutive promoter in the broad host range expression vector, pBroadgate. The regulatory effect of each cloned candidate gene was assessed by measuring the transcriptional activity of the promoterless lacZ reporter located downstream of EIL-′dapB in the genome of SBW25ΔdapB (e.g., see Fig. 1). This approach enabled regulatory effects to be determined: Negative regulators cause no increase in β-galactosidase activity in the original fusion strain, but effect significant reduction of activity in the TAF strain; positive regulators cause an increase in β-galactosidase activity in the original fusion strain, but have either no influence or effect an increase in activity in the TAF strain.
The results (Table 1) confirm regulatory connections between EIL and the elements that control their expression. In some instances the connections are already known, for example, WspF is a negative regulator of WspR (14, 15), WspR is a positive activator, via the provision of cyclic-di-GMP (16), of the wss-encoded cellulose biosynthetic cluster (17), and rspL is a positive transcriptional activator of the rsp type III secretion pathway (3, 6). In other instances, the regulators are previously uncharacterized; for example, awsX and the adjacent GGDEF motif-containing gene awsR. In four instances, two or more regulators were found to regulate a single EIL; in the case of NR9 (wssE), five different genes were implicated in transcriptional control. Two of the regulators, FleQ and AmrZ (AlgZ), are interesting, given that they have been previously assigned regulatory roles in flagella biosynthesis and alginate production (18–20). In one instance, the same regulators (CueA, CueR, CopZ) were found to affect the expression of two different EIL [NR20 (permease) and NR53 (cueA)], although the regulatory effects were reversed. CueA is the principle copper exporter that removes excess copper ions from the cytosol (14). Because the same regulators repress NR53 (the exporter) and activate NR20, they may act reciprocally, in which case NR20 could be an importer. The possibility that the NR20 locus encodes a copper uptake system awaits further investigation.
Linking EIL and Regulators to Bacterial Function.
Knowledge of positive and/or negative regulatory elements of EIL allows a connection to be made between regulators and the phenotypes they control. They also provide a means of studying the biological function of EIL and various other traits controlled by the SPyVET-identified regulators. For example, database searches revealed that cueA (identified in EIL fusion strain NR53) bears similarity to copper-transporting P-type ATPases, suggesting a role in copper homeostasis (8). This hypothesis was testable on account of the ability to overexpress cueA (by overexpression of SPyVET-identified regulator cueR; see Table 1), and confirmed experimentally because the overexpression of cueR in SBW25 increased resistance from 300 μM to 400 μM copper (data not shown). Several other phenotypic links are shown in Fig. 3. EIL fusion strain NR9, containing defects in awsX or wspF, produce the wrinkled colony morphology (Fig. 3A) typical of cellulose-overexpressing strains (14); staining with calcofluor confirmed the presence of cellulose, the product of the wss operon (17). The fleQ-defective mutant of EIL fusion strain NR9 (NR 9.5) was unable to swim through semisolid agar (Fig. 3B); it was also devoid of flagella (Fig. 3C) and was hyperresistant to H2O2-induced oxidative stress (Fig. 3D); the former is in accord with a previous study of fleQ in Pseudomonas aeruginosa (19), and the latter is previously undiscovered. Interestingly, the fleQ-mutant NR9.5 exhibited altered surface spreading (“spidery” swarming) on semisolid agar (Fig. 3C).
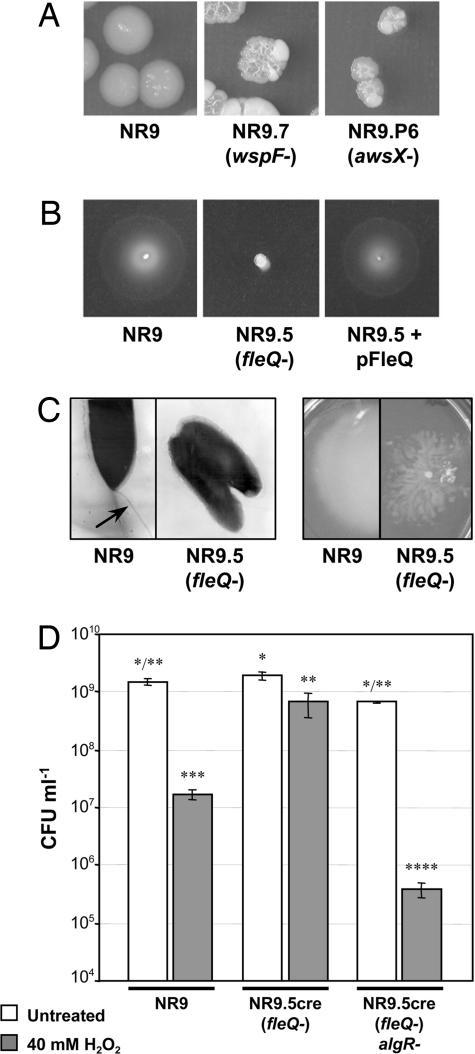
Fig. 3.
Mutations identified in TAF strains affect other phenotypes. (A) Inactivation of wspF and awsX alters colony morphology from smooth to wrinkly on M9 agar. (B and C) Inactivation of fleQ abolishes swimming motility because of a lack of flagella (B), but derepresses “spidery swarming” motility (C). (D) FleQ and AlgR control resistance to H2O2. The algR mutant displays significantly lower H2O2 resistance (P < 0.0001, d.f. = 5 by one-way ANOVA). Values are the means of three replicates, and bars are standard error of the means. Significant differences among means were revealed by Student's t test (P = 0.05) and are indicated by asterisks above the columns.
Use of SPyVET to Obtain Insight into a Regulatory Hierarchy.
SPyVET also provides opportunity for additional rounds of suppressor analysis, thus providing opportunity to define regulatory cascades. Central to this extension is the promoterless lacZ gene, which is operonic with ′dapB (see Fig. 1) and IS-Ω-Km/hah, which can be excised from the genome by Cre recombinase-mediated excision. By way of example, we show that further rounds of suppressor analysis provides insight into the regulatory relationships among the five identified regulators of wss in fusion NR9 (fleQ, amrZ, awsX, awsR, and wspF). Cre recombinase-mediated excision (SI Fig. 6) (21) was used to delete the bulk of the transposon from TAF mutants NR9fleQ and NR9wspF. The resulting strains were kanamycin-sensitive but still prototrophic, demonstrating that the 189-bp transposon scar remaining after Cre-excision (21) was sufficient for regulator inactivation. Because these modified mutants lack a repressor (and colonies turned dark blue when grown on media containing X-Gal), we reasoned that positive activators of wss transcription must be active. These positive activators could be identified by a second round of IS-Ω-Km/hah mutagenesis coupled with a screen for mutants in which lacZ was not expressed (white colonies; SI Fig. 5). For both fusion strains, ≈60,000 mutants were screened, and a single positive activator was discovered in each instance: wspR [which is negatively regulated by WspF (15, 17)] and algR (which is negatively regulated by FleQ). As described above, expression of each gene from pBroadgate in strain NR9 confirmed that each regulator activated wssE [relative to a vector-only control normalized to expression value of zero: wspR, 0.67 ± 0.29 (P = 0.0378); algR, 1.80 ± 0.13 (P < 0.0001)]. Phenotypic changes, based on prediction of regulator function, were also observed for these mutants. A defective wspR mutation (in a wspF mutant) resulted in a change of colony morphology from wrinkly to smooth [data not shown, (15)], and mutation of algR (in a fleQ mutant) increased sensitivity of SBW25 to H2O2-induced oxidative stress (22).
These two additional regulators took the total number of wss regulators to seven, of which FleQ, AmrZ, AwsX, and WspF are negative regulators, and WspR, AlgR, and AwsR are positive activators. In addition, the suppressor analysis shows that AlgR operates “downstream” of FleQ and that WspR operates downstream of WspF. Furthermore, we predict that AwsR operates downstream of AwsX, because AwsX represses wss transcription even when awsR is intact (Table 1 and Fig. 4).
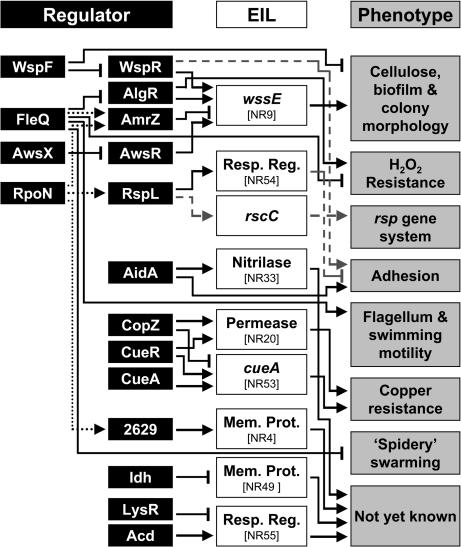
Fig. 4.
Foundation model for the molecular mechanisms underpinning the ecological success of P. fluorescens in the plant environment. Regulators (black boxes) positively activate (arrows) or repress (blunt-end lines) plant EIL (white boxes) and/or associated phenotypes (gray boxes). Interactions found in this study are shown by solid black lines, and interactions found in other studies are shown by dashed gray lines (6, 30, 39). Dotted lines are putative interactions between RpoN and potential RpoN-binding sites upstream of amrZ, rspL, and PFLU2629 (hcs). The dotted line between FleQ and AmrZ represents a putative regulatory effect based on the potential for interaction of FleQ with RpoN at the amrZ promoter; for example, FleQ (an enhancer-binding protein and positive activator of the flagellum system) may repress wss expression by activating the wss repressor AmrZ. Note that AwsX negatively regulates the wss genes and is postulated to act by repression of awsR. AidA was also found to enhance SBW25 adhesion to glass (data not shown).
Discussion
Various techniques allow the function of microbes to be studied in natural environments (23–25); however, mechanistic insight ultimately requires that the organism of interest be studied in the laboratory. Unfortunately, the act of bringing any bacterium into the laboratory means that the traits of interest may no longer be expressed. SPyVET provides a solution by virtue of the fact that it identifies regulators whose activity can be manipulated, thus enabling traits expressed in a known natural environment to be expressed in vitro (Fig. 3). Because these regulators are central to the differential expression of genes in response to different environments, they provide important information about physiological adaptation. Additionally, the ease with which subsequent rounds of suppressor analyses can proceed means that regulatory cascades otherwise hidden to experimenters can be revealed.
The regulatory connections identified by SPyVET revealed insight into the role of known regulators (Fig. 4). FleQ, AlgR, and RspL control expression of flagella (19), alginate (26), and the Rsp type III secretion pathway (6), respectively. Our findings show that these regulators also control expression of EIL, with two of them, FleQ and AlgR, playing a role in the expression of wss, and with RspL influencing the expression of a functionally uncharacterized response regulator. These data suggest a greater coordination of gene expression than realized. For example, it appears that SBW25 coordinates flagella-mediated swimming with cellulose-based surface spreading, ensuring, via FleQ, that the two modes of movement are expressed independently. Similar inverse regulation involving FleQ has been reported in P. aeruginosa; however, the key player appears to be the DNA-binding transcriptional regulator AmrZ; in a mucoid strain, AmrZ can repress fleQ (thus, flagella) while inducing transcription of the alginate gene cluster (20, 27).
In probing the function of SBW25 in its natural environment, SPyVET uncovered regulators and genes that had not been described previously (Table 1 and Fig. 4). This may reflect the fact that (for reasons of tractability) most molecular research into biological systems is constructivist; discovery is restrained within the limits of well defined and controlled in vitro conditions (28, 29). The novelty of many of the genes discovered here illustrates the power of SPyVET to reveal, en masse, new biological information about the relationship between organism and environment. Some of the previously unidentified regulators make sense in the context of the plant environment, for example, the discovery of regulators for copper transport (cueR, copZ) and nitrile metabolism (pinA) is indicative of the existence of both these compounds in the plant environment [(8); A. J. M. Howden and G. M. Preston, personal communication]. Other examples are less clear: In the case of strain NR55, the EIL encodes a putative OmpR-like regulator. Although the target of this gene is unknown, its transcription is controlled by two colocalized regulators, an activator and a repressor.
The complexity and multifactorial nature of the regulatory network controlling the SBW25 wss operon is surprising. The wss genes and their product (an acetylated cellulose polymer) (30), along with the posttranslational mechanism of cellulose enzyme activation (16, 31), have been described (14, 15, 17); however, knowledge of transcriptional control has been lacking (4). Three different regulatory pathways control wss transcription: (i) the previously known Wsp chemosensory pathway (15); (ii) FleQ, AmrZ, and AlgR; each of these regulators is known to control expression of traits other than cellulose in other organisms; and (iii) AwsXR, a previously unrecognized regulatory locus. A common feature of pathways i and iii is that they encode predicted diguanylate cyclases (DGCs) [proven in the case of WspR (16)]. This strongly implicates the secondary signaling molecule cyclic di-GMP as the ultimate effector of the activity of the wss-encoded cellulose synthases, but also highlights the fact that this molecule elicits, either directly or indirectly, marked effects at the level of transcription.
Although studying the function of bacteria in complex environments remains a significant challenge, the SPyVET strategy described here provides a straightforward genetic approach to identifying regulatory systems controlling expression of genes that often escape notice under standard laboratory conditions. Identification of these genes stands to provide insight into both the traits bacteria express in situ and the regulatory pathways that control their expression.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
Escherichia coli and P. fluorescens were routinely cultured on LB medium (32), at 37°C and 28°C, respectively. E. coli S17–1 λpir and DH5α λpir were used for maintenance, conjugation, and conjugative cloning of IVET library plasmids. For propagation, SBW25ΔdapB strains, including IVET fusion and TAF strains, were supplemented with DAP (800 μg ml−1) and lysine (80 μg ml−1), but strict auxotrophy was assessed by culturing on M9 agar and broths in the absence of DAP and lysine, where auxotrophs displayed no detectable growth after 48 h. Gentamycin (Gm; 10 μg ml−1), kanamycin (Km; 25 μg ml−1), tetracycline (Tc; 10 μg ml−1), CFC (0.5 × strength; Oxoid, Hampshire, U.K.), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg ml−1) were used in growth media, as appropriate. Vector pCre (21) was used for Cre-loxP-mediated deletion of IS-Ω-Km/hah by introducing pCre transiently into TAF strains and selecting for colonies that were TcR and KmS.
Construction of IS-Ω-Km/hah, Mutagenesis, and Reversion Frequencies.
Plasmid pSCR001 (10571 bp; GenBank accession no. DQ059989) carrying IS-Ω-Km/hah (SI Fig. 6) was constructed by replacing the CmR gene and the section of phoA from IS-phoA/hah (33) with Omegon-Km (Ω-Km) from pJFF350 (34); cloning details are available upon request. IS-Ω-Km/hah includes the IE (and nptII promoter), OE, and loxP sites of IS-phoA/hah and the KmR gene and ColE1 origin of replication of Ω-Km bordered by transcriptional and translational stop sequences at each end (SI Fig. 6). Mutation of IVET strains using IS-Ω-Km/hah and miniTn5Km was performed essentially as described (12). Mutants were plated on M9 agar with Tc, Km, and X-Gal to identify TAF strains, and prototrophy of TAF strains was further confirmed by restreaking on fresh M9 agar in the absence of DAP and lysine. Background reversion frequencies for each recipient (IVET fusion) strain were determined by enumerating prototrophic colonies arising from 10-fold serial dilutions of an overnight broth of each strain spread on M9+Tc agar.
Molecular Biology.
Arbitrary primed (AP)-PCR and sequencing (13) was used to identify the genomic location of IS-Ω-Km/hah transposon insertions by using primers designed by Manoil (21). To locate miniTn5Km insertions, primers to the O-end of the transposon were designed for AP-PCR: round 1, miniTn5F3 5′-ATCGGGCCTTGATGTTACCGAG; round 2 and sequencing, miniTn5F4, 5′-TACCCAGTCTGTGTGAGCAGG.
Transposon insertions in regulatory loci were confirmed by PCR using primers that flank the insertion (available upon request). Regulatory gene candidates were PCR amplified by using Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA) and KOD polymerase (Novagen, San Diego, CA) to include only the predicted Shine–Dalgarno site, stop and start codons, and some downstream sequence (primers available upon request). Regulatory gene candidates were cloned and constitutively expressed by using Gateway technology (Invitrogen) employing the entry vector pENTR/D-TOPO (Invitrogen) and broad host range destination vector pBroadgate (a kind gift from R. Thwaites and J. Mansfield, Imperial College, London). Fidelity of cloned PCR products was verified by DNA sequencing. A plasmid control for use in expression studies, pBroadgate-D, was constructed by replacing the ccdB gene in pBroadgate with a small noncoding DNA fragment, 5′-TCTAGCTAGTTATTCAGGTG-3′.
β-Galactosidase Assays.
The transcriptional activity of IVET fusions was assessed by using the promoterless reporter gene lacZ that is located downstream of ′dapB in pIVETD (4). β-galactosidase activity was assayed by monitoring the hydrolysis of 4-methylumbelliferyl-β-d-galactoside to yield the fluorescent product, 7-hydroxy-4-methylumbelliferone (4MU) as described (Amersham Pharmacia, Piscataway, NJ). The 4MU was detected at 460 nm after excitation at 365 nm using a Polarstar plate reader (BMG Labtech, Aylesbury, U.K.). Optical densities of cultures were recorded, and enzyme activity was normalized to the cell density. Data were subject to one-way ANOVA by using JMP software version 5.1 (SAS Institute, Cary, NC).
Bioinformatic Analyses.
The library inserts of the 27 IVET fusions tested were delimited by sequencing the cloning junctions of pIVETD fusion constructs that were recovered from fusion strains by conjugative cloning (35). These sequence data and transposon insertion locations were mapped to the SBW25 genome by using Artemis V7 software (36). Circular chromosomal maps were created in XML language by using CGView (37). Putative promoters were identified by PROMSCAN (http://molbiol-tools.ca/promscan/).
Phenotypic Analyses.
To determine the presence of flagella, P. fluorescens cells were negatively stained on Formvar/Carbon-coated copper grids with 2% uranyl acetate for 2 min and then soaked in water for 10 min before observation with a CM10 TEM (Philips, Eindhoven, The Netherlands). The motility of P. fluorescens strains in agar was assessed by stabbing a sterile wire needle into an overnight broth culture and then into the center of a 0.25% LB agar plate. Motility was assessed after 12 h. Colony morphologies of wspF and awsX mutants were determined 96 h after streaking bacterial strains to single colonies on M9 agar with appropriate supplements. Copper resistance was assessed by determining the minimal inhibitory concentration (MIC) of copper sulfate in M9 agar plates with DAP and lysine. The MIC value was considered the lowest concentration that inhibits the formation of single colonies after 3 days incubation at 28°C. Resistance to oxidative damage was assessed by exposing cells to 40 mM hydrogen peroxide for 15 min and enumerating survivors (38).
Supplementary Material
Acknowledgments
We thank Christopher Knight for help with statistical analyses and critical reading of the manuscript; John Baker, Julie Bull, Zena Robinson, and Julian Robinson for technical help; and David Patton for electron microscopy. This work was supported by European Union Grant QLRT-2001-00914 (Pseudomics) (to S.R.G.), Biotechnology and Biological Sciences Research Council (U.K.) (C.D.M. and R.W.J.), and the Gatsby Charitable Foundation (S.M.G.).
Abbreviations
- DAP
diaminopimelic acid
- EIL
environment-induced loci
- IVET
in vivo expression technology
- SPyVET
suppressor IVET
- TAF
transcriptionally activated fusion.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ059989).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706739104/DC1.
References
- 1.Mahan MJ, Slauch JM, Mekalanos JJ. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 2.Rainey PB. Environ Microbiol. 1999;1:243–257. doi: 10.1046/j.1462-2920.1999.00040.x. [DOI] [PubMed] [Google Scholar]
- 3.Preston GM, Bertrand N, Rainey PB. Mol Microbiol. 2001;41:999–1014. doi: 10.1046/j.1365-2958.2001.02560.x. [DOI] [PubMed] [Google Scholar]
- 4.Gal M, Preston GM, Massey RC, Spiers AJ, Rainey PB. Mol Ecol. 2003;12:3109–3121. doi: 10.1046/j.1365-294x.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X-X, Lilley A, Bailey MJ, Rainey PB. FEMS Microbiol Ecol. 2004;51:9–17. doi: 10.1016/j.femsec.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Jackson RW, Preston GM, Rainey PB. J Bacteriol. 2005;187:8477–8488. doi: 10.1128/JB.187.24.8477-8488.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X-X, George A, Bailey MJ, Rainey PB. Microbiology. 2006;152:1867–1875. doi: 10.1099/mic.0.28731-0. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X-X, Rainey PB. Mol Plant–Microbe Interact. 2007;20:581–588. doi: 10.1094/MPMI-20-5-0581. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X-X, Rainey PB. Genetics. 2007;176:2165–2176. doi: 10.1534/genetics.107.075713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson RW, Giddens SR. Infect Disord Drug Targets. 2006;6:207–240. doi: 10.2174/187152606778249944. [DOI] [PubMed] [Google Scholar]
- 11.Rediers H, Rainey PB, Vanderleyden J, Mot R. Microbiol Mol Biol Rev. 2005;269:217–261. doi: 10.1128/MMBR.69.2.217-261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, et al. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goymer P, Kahn SG, Malone JG, Gehrig SM, Spiers AJ, Rainey PB. Genetics. 2006;173:515–526. doi: 10.1534/genetics.106.055863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bantinaki E, Kassen R, Knight CG, Robinson Z, Spiers AJ, Rainey PB. Genetics. 2007;176:441–453. doi: 10.1534/genetics.106.069906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malone JG, Williams R, Christen M, Jenal U, Spiers AJ, Rainey PB. Microbiology. 2007;153:980–994. doi: 10.1099/mic.0.2006/002824-0. [DOI] [PubMed] [Google Scholar]
- 17.Spiers AJ, Kahn SG, Bohannon J, Travisano M, Rainey PB. Genetics. 2002;161:33–46. doi: 10.1093/genetics/161.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baynham PJ, Wozniak DJ. Mol Microbiol. 1996;22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 19.Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tart AH, Blanks MT, Wozniak DJ. J Bacteriol. 2006;188:6483–6489. doi: 10.1128/JB.00636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manoil C. Methods Enzymol. 2000;326:35–47. doi: 10.1016/s0076-6879(00)26045-x. [DOI] [PubMed] [Google Scholar]
- 22.Lizewski SE, Schurr JR, Jackson DW, Frisk A, Carterson AJ, Schurr MJ. J Bacteriol. 2004;186:5672–5684. doi: 10.1128/JB.186.17.5672-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumont MG, Murrell JC. Nat Rev Microbiol. 2005;3:499–504. doi: 10.1038/nrmicro1162. [DOI] [PubMed] [Google Scholar]
- 24.Pernthaler J, Amann RI. Microbiol Mol Biol Rev. 2005;69:440–461. doi: 10.1128/MMBR.69.3.440-461.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner M, Nielsen PH, Loy A, Nielsen JL, Daims H. Curr Opin Biotechnol. 2006;17:83–91. doi: 10.1016/j.copbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Deretic V, Gill JF, Chakrabarty AM. Nucleic Acids Res. 1987;15:4567–4581. doi: 10.1093/nar/15.11.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baynham PJ, Brown AL, Hall LL, Wozniak DJ. Mol Microbiol. 1999;33:1069–1080. doi: 10.1046/j.1365-2958.1999.01550.x. [DOI] [PubMed] [Google Scholar]
- 28.Roll-Hansen N. Stud Hist Philos Sci C. 1998;29:165–187. [Google Scholar]
- 29.Kassen R, Rainey PB. Annu Rev Microbiol. 2004;58:207–231. doi: 10.1146/annurev.micro.58.030603.123654. [DOI] [PubMed] [Google Scholar]
- 30.Spiers AJ, Bohannon J, Gehrig SM, Rainey PB. Mol Microbiol. 2003;50:15–27. doi: 10.1046/j.1365-2958.2003.03670.x. [DOI] [PubMed] [Google Scholar]
- 31.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 33.Bailey J, Manoil C. Nat Biotechnol. 2002;20:839–842. doi: 10.1038/nbt715. [DOI] [PubMed] [Google Scholar]
- 34.Fellay R, Krisch HM, Prentki P, Frey J. Gene. 1989;76:215–226. doi: 10.1016/0378-1119(89)90162-5. [DOI] [PubMed] [Google Scholar]
- 35.Rainey PB, Heithoff DM, Mahan MJ. Mol Gen Genet. 1997;256:84–87. doi: 10.1007/s004380050548. [DOI] [PubMed] [Google Scholar]
- 36.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 37.Stothard P, Wishart DS. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 38.Ma JF, Ochsner UA, Klotz MG, Nanayakkara VK, Howell ML, Johnson Z, Posey JE, Vasil ML, Monaco JJ, Hassett DJ. J Bacteriol. 1999;181:3730–3742. doi: 10.1128/jb.181.12.3730-3742.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulesekara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, et al. Proc Natl Acad Sci USA. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.