Abstract
Caspase-like proteases are universal mediators of programmed cell death (PCD). Because plants have no caspase homologs, establishing the nature of their caspase-like activities is of considerable importance to our understanding of PCD in plants. Caspase-3, displaying DEVD specificity, is a key executioner caspase in animal cells. Self-incompatibility (SI) is an important mechanism to prevent self-fertilization and inbreeding in higher plants by inhibiting incompatible pollen. In Papaver rhoeas, SI activates a caspase-3-like/DEVDase activity in incompatible pollen that plays a pivotal role in regulating PCD. Here we characterize the SI-induced caspase-like activities in detail; our work provides insights into the temporal and spatial activation of plant caspase-like enzymes. We show that SI also activates a VEIDase and a LEVDase and that the VEIDase plays a role in SI-induced PCD. The DEVDase and VEIDase are activated remarkably rapidly: detectable within 1–2 h after SI induction; the LEVDase activity peaks later. Importantly, we show live-cell imaging of a DEVDase activity in a higher plant cell; the SI-activated DEVDase has a cytosolic and nuclear localization. We also demonstrate that SI induces a rapid and substantial cytosolic acidification that matches the in vitro pH optima for the SI-induced caspase activities. Because both cytosolic acidification and nuclear caspase localization are observed during apoptosis in animal cells, our data provide striking parallels between SI-induced PCD and apoptosis in animal cells.
Keywords: caspase–like activity, programmed cell death, acidification
Apoptosis or programmed cell death (PCD) is an important, highly conserved process used to remove unwanted eukaryotic cells. It involves the highly regulated death of targeted cells during development and defense against pathogens. In animal cells, caspases, cysteine aspartate-specific proteases, are key players involved in mediating initiation and execution of cell death. Caspases are highly specific endopeptidases that, upon activation by proteolytic processing, cleave target substrates after an aspartic acid residue. Use of specific peptide substrates established the substrate specificities of animal caspases (1). Caspase-3, with the optimal tetrapeptide recognition motif DEVD, together with caspase-6 (VEID), is the key “executioner” caspase. The initiator caspases-8 and -9 preferentially recognize (L/V)EXD motifs.
Because yeast and plants have no caspase gene homologs (2), the nature of their caspase-like enzymes is currently the subject of considerable debate. Metacaspases that structurally resemble animal caspases have been identified in fungi, protozoa, and plants (3–5). However, although data implicate their involvement in PCD (5–7), biochemical characterization shows that metacaspases do not cleave caspase-specific substrates; many are arginine/lysine-specific proteases (7).
Despite this conundrum, considerable biochemical evidence exists for the involvement of caspase-like activities in PCD in plants (8). Use of synthetic inhibitor peptides and fluorogenic substrates, based on the caspase optimal tetrapeptide recognition motifs, has allowed identification of several caspase-like activities in plants. For example, a plant VEIDase (caspase-6) is activated during embryo development (6, 9). Vacuolar processing enzyme has YVADase (caspase-1) activity and functions in PCD in the hypersensitive response to plant pathogens (10, 11). Plant “saspases” (serine aspartate-specific proteases) and a TATDase have also been identified (4, 12). Although these enzymes are probably functionally equivalent to caspases, they are not caspase-3-like/DEVDase proteases. The identification of a DEVDase activity in several higher plants is of considerable interest because it suggests that a protein with characteristics similar to a mammalian caspase-3 activity exists and mediates PCD. It has been identified in cell extracts through cleavage of a caspase-3-specific substrate, Ac-DEVD-7-amino-4-trifluoromethyl coumarin (AMC) and use of DEVD tetrapeptide inhibitors that alleviate PCD (13, 14). Most display both DEVDase and YVADase activities (13, 15, 16); however, self-incompatibility (SI) in Papaver involves a DEVDase and not a YVADase activity (17).
SI is an important mechanism used to prevent self-fertilization in flowering plants. “Self” (incompatible) pollen is “recognized” and rejected, whereas “cross” (compatible) pollen is allowed to grow. Use of a multiallelic S locus allows S-specific pollen recognition by the pistil through interaction of pollen and pistil S determinants with matching alleles. Several different SI systems exist, with quite distinct molecular and genetic control (18, 19). In Papaver rhoeas, the field poppy, the pistil S locus determinants (S proteins) are small ≈15-kDa proteins (20). They act as ligands, triggering a Ca2+- dependent signaling network in incompatible pollen (21). SI can be triggered by the addition of recombinant pistil S proteins to pollen tubes growing in vitro, allowing analysis of events triggered specifically in incompatible pollen. Early events are involved in the rapid arrest of pollen tube growth, including depolymerization of the F-actin cytoskeleton (22) and Ca2+/phosphorylation-mediated inhibition of soluble inorganic pyrophosphatase activities (23). Actin depolymerization and activation of a MAPK, p56, mediate PCD-related events (14, 24), including cytochrome c leakage into the cytosol, DNA fragmentation, and activation of a caspase-3-like/DEVDase activity (17). This DEVDase plays a key functional role in pollen tube PCD (14, 17).
Here we provide detailed characterization of the caspase-like activities stimulated by SI in incompatible pollen. Three distinct activities, a DEVDase, VEIDase, and a LEVDase, were identified. The former two are activated surprisingly rapidly, which was accompanied by dramatic cytosolic acidification. Moreover, the DEVDase activity has a cytosolic and nuclear localization.
Results
Visualization of SI-Induced DEVDase Activities in Pollen Tubes.
Caspase-3 activities can be measured in living cells with probes based on a substrate containing the DEVD sequence. These substrates are cleaved by activated DEVDases, releasing a fluorescent cleavage product, allowing measurement of DEVDase activity in vivo. Having previously identified a SI-stimulated DEVDase, we wished to investigate its intracellular localization in living pollen tubes.
The NucView 488 live-cell probe, when cleaved by a DEVDase, releases a DNA-binding dye that migrates to the nucleus, fluorescing upon binding to DNA. Thus, this probe reports intracellular activated DEVDase but not its localization. We used this probe to verify the specificity of the SI-induced DEVDase activity in incompatible pollen tubes. Few untreated pollen tubes exhibited DEVDase activity (8 ± 0%), whereas SI stimulated a high incidence of this activity (89 ± 9%) (Fig. 1a). Pretreatment of pollen tubes with the tetrapeptide caspase inhibitor Ac-DEVD-CHO significantly reduced the incidence of SI-induced DEVDase activity to 24 ± 11% (***, P < 0.001). In contrast, pollen tubes pretreated with Ac-YVAD-CHO had a level of DEVDase activity similar to that of SI-treated (75 ± 16%) (Fig. 1a). DEVDase activity is reported with tight specificity in live cells because these data show the same specificity exhibited by our previous study (17).
Fig. 1.
Live-cell DEVDase activity in pollen tubes. (a) Specificity of DEVDase in SI-stimulated pollen tubes. Pollen tubes were scored for DEVDase activity by using the NucView probe 5 h after SI induction. Fluorescent nuclei indicated high caspase activity (+, filled bars); low fluorescence indicated no caspase activity (−, open bars); intermediate fluorescence, intermediate activity (+/−, cross-hatched bars). UT, untreated; +DEVD, pretreatment with Ac-DEVD-CHO; +YVAD, pretreatment with Ac-YVAD-CHO. Error bars, ± SEM (n = 3). (b–j) CR(DEVD)2 imaging of DEVDase activity in SI-induced pollen. (b) CR-(DEVD)2 live-cell imaging of a normally growing untreated pollen tube (control). No background DEVDase activity was detected. (c) Bright-field image of b. (d) DAPI staining shows nuclear DNA in the GC and VN. (e) Typical CR fluorescence upon SI induction. DEVDase activity is cytosolic and also localizes with the GC and VN. (f) CR fluorescence 2 h after SI induction. (g) Bright-field image of pollen tube in f. Note the highly granular cytoplasm. (h) CR fluorescence 4 h after SI induction. (i) Typical example of CR fluorescence in a pollen grain 4 h after SI induction. (j) No DEVDase activity is detected in an untreated pollen grain at 4 h. Images shown are median plane confocal sections; all were captured using identical detection settings. (Scale bars: 10 μm.)
Having verified the authenticity of the DEVDase activity detected in live cells, we examined the localization of DEVDase activity in living cells by using CR(DEVD)2. This probe uses the fluorophore cresyl violet (CR) coupled to two DEVD peptides. When a DEVDase cleaves DEVD-CR linkages, the fluorescent CR marker is released, providing temporal, spatial, and quantitative information about the activated DEVDase. Fig. 1b shows a typical unstimulated pollen tube, with no fluorescence, and corresponding bright-field image (Fig. 1c). Pollen tubes have a vegetative nucleus (VN), and a generative cell (GC) nucleus, visualized with DAPI staining (Fig. 1d). In SI-induced pollen tubes, CR fluorescence revealed DEVDase activity in the cytosol, VN, and GC (Fig. 1 e, f, and h). The cytoplasm was more granular in appearance (Fig. 1g). Notably, the GC exhibited high DEVDase activity, suggesting that it is preferentially targeted. The same localization of DEVDase activity was exhibited by pollen grains after SI induction (Fig. 1i), whereas no activity could be detected in untreated pollen (Fig. 1j).
SI-Induced Pollen Exhibits Several Caspase-Like Activities.
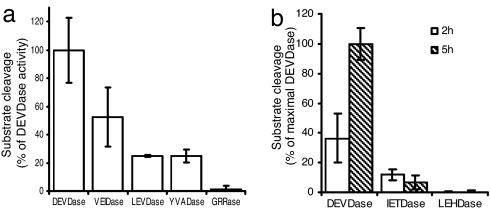
We previously showed, by using DNA fragmentation combined with caspase inhibitors, that SI involves a DEVDase but not a YVADase (17). Here, the use of AMC-based peptide caspase substrates that act as fluorogenic indicators for caspase activities enabled us to characterize the SI-induced caspase-like activities more directly and in more detail in pollen extracts. Fig. 2a shows that the substrates Ac-DEVD-AMC (caspase-3) and Ac-VEID-AMC (caspase-6) are cleaved by proteases activated in SI-induced pollen. The DEVDase exhibited 52.3 ± 21% higher activity than the VEIDase. Ac-LEVD-AMC (caspase-4) and Ac-YVAD-AMC (caspase-1) were also cleaved but at much lower levels (24.9 ± 1% and 24.9 ± 5% that of DEVDase levels). This finding provides good evidence that SI induces executioner caspase activities.
Fig. 2.
Substrate specificity of SI-induced caspase-like protease activities. (a) SI-induced pollen extracts were tested for substrate specificity by using different caspase substrates: Ac-DEVD-AMC (DEVDase), Ac-VEID-AMC (VEIDase), Ac-LEVD-AMC (LEVDase), Ac-YVAD-AMC (YVADase), Ac-GRR-MCA (GRRase). Relative fluorescence units were measured and are expressed as a percentage of the DEVDase activity. Error bars, ± SEM (n = 3). (b) Initiator caspase activities are not detected in SI-induced samples. The initiator caspase substrates Ac-IETD-AMC and Ac-LEHD-AMC show low or no cleavage activities at 2 h and 5 h after SI induction. Error bars, ± SEM (n = 3).
To examine whether a metacaspase activity, identified in several plant systems (7, 25), was stimulated by SI we tested for cleavage of the substrate Ac-GRR-4-methyl-coumaryl-7-amide (MCA). No GRRase activity (1.4 ± 2%) was induced (Fig. 2a). There was also no evidence that SI triggers “initiator” caspase-like activities. SI-induced pollen extracts at 2 h and 5 h displayed very little cleavage of Ac-IETD-AMC (caspase-8), and there was no cleavage of Ac-LEHD-AMC (caspase-9) (Fig. 2b).
Establishing SI-Induced Caspase Substrate Specificities.
We characterized the SI-induced DEVDase, VEIDase, and LEVDase caspase-like activities further, but not the YVADase activity, because it was low, even 8 h after SI (unpublished data), and we had shown that it was not involved in SI (17). Fig. 3a shows Ac-DEVD-AMC, Ac-VEID-AMC, and Ac-LEVD-AMC substrate cleavage activities in the presence of a range of tetrapeptide caspase-specific inhibitors. As expected from previous studies (17)Ac-YVAD-CHO did not inhibit any of these activities. Surprisingly, the pan-caspase inhibitor Ac-VAD-CHO was also not effective against these activities (Fig. 3a). Although the SI-induced DEVDase was inhibited by Ac-DEVD-CHO, the VEIDase and LEVDase activities were also inhibited. Taking the untreated control as 100% activity, Ac-DEVD-CHO significantly inhibited the SI-induced DEVDase activity by 71 ± 1% (***, P < 0.001), the SI-induced VEIDase activity by 77 ± 1% (P < 0.001), and LEVDase by 46 ± 3% (P = 0.002). Ac-VEID-CHO significantly inhibited the SI-induced VEIDase by 75 ± 2% (P < 0.001) and also inhibited the DEVDase and LEVDase activities by 81 ± 2% (P < 0.001) and 57 ± 3% (P < 0.001), respectively. Furthermore, besides inhibiting its corresponding LEVDase activity, Ac-LEVD-CHO inhibited both the DEVDase and VEIDase by 65% (P < 0.001). The similar sensitivities of the DEVDase, VEIDase, and LEVDase activities suggested that they may be related; however, other data (see later) indicate they probably represent separate activities. Other tetrapeptide inhibitors, Ac-IETD-CHO (caspase-8), Ac-LEHD-CHO (caspase-9), and Ac-IEPD-CHO (granzyme B) did not significantly inhibit the SI-induced activities (Fig. 3a). Although interpretation of the exact caspase specificities should be viewed with caution, the use of these caspase-specific peptides is useful for characterization, providing information about the recognition sites involved. The data suggest that SI induces three caspase-like activities: a DEVDase, a VEIDase, and a LEVDase.
Fig. 3.
Effect of caspase inhibitors on SI-induced caspase-like activities and DNA fragmentation. (a) Ac-DEVD-AMC (filled bars), Ac-VEID-AMC (open bars), and Ac-LEVD-AMC (cross-hatched bars) substrate cleavage in the presence of caspase-specific tetrapeptide inhibitors. Substrate cleavage 5 h after SI induction is expressed as a percentage of activity in the presence of DMSO (100%). Error bars, ± SEM (n = 3). (b) DNA fragmentation in untreated (UT), SI-induced (SI), and in SI-induced after pretreatment with Ac-DEVD-CHO (SI+DEVD), Ac-VEID-CHO (SI+VEID), or Ac-LEVD-CHO (SI+LEVD) inhibitors before SI induction. Error bars, ± SEM (n = 300).
To establish whether the VEIDase or LEVDase is involved in mediating SI-induced PCD, we tested whether pretreatment with the inhibitor Ac-VEID-CHO or Ac-LEVD-CHO before SI alleviated PCD, using DNA fragmentation to measure it (Fig. 3b). SI-induced DNA fragmentation was significantly inhibited by Ac-DEVD-CHO (**, P = 0.002) and Ac-VEID-CHO (*, P = 0.0212), but not by Ac-LEVD-CHO (P = 0.314, not significant; n = 300 in each case). This result provides good evidence that both the DEVDase and VEIDase are functionally involved in SI-mediated PCD.
Most caspase-like activities are insensitive to broad-spectrum cysteine and serine protease inhibitors (6, 9, 10, 13, 15, 16). However, some inhibitors, notably leupeptin (7), inhibit metacaspases. The SI-induced caspase-like activities were not inhibited by PMSF, aprotinin, pepstatin, leupeptin, 7-amino-1-chloro-3-tosylamido-2-heptanone, and E-64 at their maximal recommended concentrations [supporting information (SI) Table 2], confirming that SI activates authentic caspase-like activities.
Temporal Activation of the SI-Induced Caspase Activities.
We examined the temporal activation of the three SI-induced caspase-like activities in pollen extracts by measuring their cleavage of tetrapeptide-AMC substrates. A small increase in DEVDase activity (P = 0.568, not significant) was detected 90 min after SI induction; significantly increased activity (72.3%; *, P = 0.041) was detected at 3 h; this activity peaked 5 h after SI induction (**, P = 0.006; Fig. 4a). The temporal profile of the VEIDase activity was very similar: increased at 3 h (*, P = 0.049) and peaking at 5 h (*, P = 0.026). Although the LEVDase activity was only 40.4% and 42.9% of the DEVDase activity at 3 h and 5 h, respectively, the increases were significant (***, P < 0.001). Furthermore, its activity was still increasing at 8 h, when the DEVDase and VEIDase activities had significantly decreased to 44.6% and 28.5% of their peak levels at 5 h, respectively. Eight hours after SI induction, the LEVDase activity was 52% higher than that of the DEVDase. These data clearly show that DEVDase and VEIDase activities are stimulated early in SI-stimulated PCD, whereas the LEVDase activity peaks later.
Fig. 4.
Temporal activation of SI-induced caspase-like activities. (a) SI-induced caspase activities were followed temporally in pollen extracts after SI induction, by using the Ac-DEVD-AMC (♦), Ac-VEID-AMC (○), and Ac-LEVD-AMC (●) substrates. Error bars, ± SEM (n = 3). (b) DEVDase activity in pollen tubes measured with the NucView probe. The number of pollen tubes with highly fluorescent nuclei (+, filled bars), little to no fluorescent nuclei (−, open bars), and nuclei with intermediate fluorescence (+/−, hatched bars) was scored in untreated (UT) and SI-induced (SI) pollen tubes. Fluorescence intensity indicates the level of DEVDase activity. Error bars, ± SEM (n = 3). (c) Early DEVDase activity visualized in a pollen tube with the CR(DEVD)2 probe 44, 59, and 95 min after SI induction. (Scale bar: 10 μm.) (d) Example of cytosolic and nuclear DEVDase increases detected at 108 and 190 min after SI using CR(DEVD)2. (Scale bar, 10 μm.)
Quantitative analysis of the temporal activation of the DEVDase activity in live cells revealed that pollen tubes exhibiting DEVDase activity were rarely observed at 1 or 2 h after SI (Fig. 4b). However, many pollen tubes (57.2%) showed DEVDase activity 3 h after SI induction, indicating rapid activation between 2 and 3 h. Most pollen tubes (89.2%) exhibited DEVDase activity at 5 h. Live-cell imaging revealed that cytosolic DEVDase activity could be detected as early as 1–2 h in some pollen tubes (Fig. 4c). Increasing DEVDase activity in the VN and GC was visualized later (Fig. 4d). Together, these data reveal early and rapid activation of SI-induced DEVDase in pollen tubes.
SI-Induced Caspase-Like Activities Have an Acidic pH Optimum.
Because pH can affect enzyme activity, we ascertained the optimal pH for the SI-induced caspase-like activities, which revealed that the pH range for the DEVDase and VEIDase is very narrow, with peak activity at pH 5.0; activities were sharply reduced at other pH levels (Fig. 5a). The LEVDase was slightly less pH-sensitive, but its optimum was still acidic (pH 4–6). The DEVDase and VEIDase exhibited different maximal activities depending on buffer composition (SI Fig. 6), suggesting that they require different conditions for optimal activity.
Fig. 5.
SI-induced caspase-like activities are optimal at an acidic pH, and cytosolic acidification is triggered during SI. (a) Ac-DEVD-AMC (♦), Ac-VEID-AMC (○), and Ac-LEVD-AMC (●) substrate cleavage activities at different pH values, with citrate/phosphate buffers, 5 h after SI induction. Activity is expressed as percentage of maximal activity at pH 5.0. Error bars, ± SEM (n = 3). (b) SI-induced [pH]i changes measured in individual pollen tubes 1–4 h after SI induction. Error bars, ± SEM (n = 13 and 12).
SI Results in Intracellular Acidification.
We investigated whether SI resulted in cytosol acidification because it is observed during apoptosis in animal cells (26, 27), and the pH optima of the SI-induced caspase-like activities were unusually acidic. Measurement of the intracellular cytosolic pH ([pH]i) with the pH-sensitive fluorophore 2′-7′-bis(carboxyethyl)-5(6)-carboxyfluorescein (BCECF) in normally growing and SI-induced pollen tubes revealed that the [pH]i is dramatically altered after SI (Fig. 5b). Normally growing pollen tubes exhibited a [pH]i of 6.9 ± 0.04 (n = 13), whereas SI-induced pollen tubes had a [pH]i of 5.5 ± 0.08 (n = 12). The cytosolic acidification observed fits the pH optima for the SI-induced caspase activities very closely.
DEVDase and VEIDase Activities Have Different Sensitivities to Cations.
Because the SI-induced DEVDase and VEIDase had similar temporal activation profiles and pH optima, we wondered whether they represented the same activity. LEVD was clearly different because of its later temporal activation. Animal caspases are sensitive to certain cations, e.g., Zn2+ (28), so we tested the SI-induced DEVDase and VEIDase sensitivities to cations to see whether they had different properties.
The SI-activated caspase-like activities were sensitive to 100 mM Zn2+; DEVDase and VEIDase activities lost 94% and 82% of their activities, respectively (***, P = 0.001; Table 1). The DEVDase was inhibited by 100 mM Ca2+ (43%; ***, P < 0.001), whereas the VEIDase activity was less sensitive (21% inhibition). Similarly, 100 mM MgCl2 inhibited the DEVDase activity by 45%, whereas the VEIDase was only inhibited by 14%. Both the DEVDase and VEIDase were highly sensitive to 500 mM NaCl and 500 mM KCl (both ***, P < 0.001), but the DEVDase was much more sensitive than the VEIDase activity (**, P = 0.004 and *, P = 0.034, respectively; Table 1). These differences in ion sensitivity suggest that the DEVDase and VEIDase are distinct.
Table 1.
Effect of cations on DEVDase and VEIDase activities
| Salt | Activity, % of control |
|
|---|---|---|
| DEVDase | VEIDase | |
| ZnCl2 | ||
| 0.1 mM | 101 ± 5 | 117 ± 2 |
| 1 mM | 93 ± 5 | 112 ± 3 |
| 10 mM | 88 ± 5 | 120 ± 6 |
| 100 mM | 6 ± 5 | 18 ± 1 |
| CaCl2 | ||
| 1 mM | 100 ± 1 | 108 ± 5 |
| 10 mM | 98 ± 2 | 104 ± 5 |
| 100 mM | 57 ± 1 | 79 ± 4 |
| MgCl2 | ||
| 10 mM | 95 ± 4 | 102 ± 4 |
| 100 mM | 55 ± 3 | 86 ± 1 |
| 500 mM | 53 ± 6 | 21 ± 1 |
| NaCl | ||
| 10 mM | 99 ± 1 | 94 ± 3 |
| 100 mM | 80 ± 5 | 86 ± 2 |
| 500 mM | 40 ± 3 | 78 ± 3 |
| KCl | ||
| 10 mM | 100 ± 2 | 104 ± 5 |
| 100 mM | 80 ± 4 | 86 ± 3 |
| 500 mM | 38 ± 3 | 69 ± 5 |
Activities were measured using Ac-DEVD-AMC and Ac-VEID-AMC fluorescent caspase peptide substrates. Data are the mean of three independent experiments. Error bars are ± SEM.
Discussion
Visualization of SI-Induced Caspase-3-Like/DEVDase Activity in Pollen Tubes.
Because DEVDases are key mediators of the execution of apoptosis/PCD in eukaryotes, imaging their activity provides important spatial and temporal data. Few studies visualizing caspase-like activities in higher plant cells exist, and none shows a DEVDase activity. A VEIDase was detected in developing barley endosperm (9); a generic caspase inhibitor, FITC-VAD-fmk, visualized caspase-like activity in the cytosol and nucleus during PCD in tobacco cell cultures (29), but the specificity was unknown. Because the SI-induced DEVDase activity is not inhibited by VAD, it must represent a different enzyme. The only other existing study was in Chara (an alga), but fluorescence was not elevated, and the cells did not die (16). Our studies show that SI induces DEVDase activation in living pollen tubes, and they image DEVDase in response to a physiologically relevant stimulus in a plant cell.
The cytosolic and nuclear localization of the SI-induced DEVDase activity is of considerable interest. It has been observed in a variety of animal cells, and it is well established that activated caspase-3 translocates from the cytosol to the nucleus during apoptosis (30, 31). Our work provides important spatial information suggesting further parallels between animal and plant caspases. Targeting of the GC nucleus provides a way to kill incompatible pollen. The timing of elevated SI-induced DEVDase activity, coupled with its nuclear localization, fits well with earlier observations of DNA fragmentation initially being detected 4 h after SI (32). Thus, the SI-induced DEVDase is activated in the correct localization shortly before its presumed effects are first detected.
Temporal Activation of the SI-Induced Caspase-Like Activities.
Our data suggest that the DEVDase and VEIDase are involved in SI-mediated PCD. The activation of both of these activities is rapid. DEVDase activity was observed as early as 1 h after SI, and substantial numbers of pollen tubes exhibited DEVDase activity ≈3 h after SI induction. This finding is consistent with analysis of animal caspase-3, which, once initiated, is rapidly activated (33). The LEVDase exhibited a different temporal activation profile; although activity was low initially, activity was still increasing at 8 h. Although data show that Ac-LEVD-CHO does not inhibit DNA fragmentation, we suggest that the LEVDase may play a role in later stages of PCD, downstream of the DEVDase and VEIDase.
Importantly, the caspase-like activities observed are induced by a specific, biologically relevant ligand, rather than a nonspecific stress or drug. This finding represents, to our knowledge, the most rapid example of caspase-like activation in a plant cell by a physiologically relevant stimulus. Few studies have examined the temporal activation of caspase-like activities in plant cells, and in existing examples, activities are often measured in days rather than hours. For example, the VEIDase in pine embryo was maximally activated at 4 days (6, 9). Heat shock of BY-2 cells gave maximal DEVDase activity at 15 h (34). Although a DEVDase was rapidly activated in Arabidopsis protoplasts 30 min after UV-C radiation (13), this trigger is not physiologically relevant to plants. Interestingly, when we examined activation of the DEVDase-like activity by mastoparan, it was activated far more rapidly, within 30 min (SI Fig. 7), indicating that artificial stimulation of PCD does not give an accurate picture of temporal events.
SI Caspase-Like Activities Are Distinct from Others Identified in Plants.
SI activates three caspase-like activities. We discounted the low YVADase activity because we showed previously that it was not involved in SI-mediated PCD (17). Identification of a DEVDase confirms previous data, based on DNA fragmentation and caspase inhibitors (14, 17). We show that SI-induced PCD is also mediated by a VEIDase activity. The high LEVDase activity, induced later than the DEVDase and VEIDase, suggests that it may function in later cellular dismantling events. The plant caspase-like genes cloned to date are a YVADase (10, 11, 35) and metacaspases (7, 25). Several caspase-like activities have been identified, for example, a UV-light stimulated DEVDase in Arabidopsis (13) and a VEIDase during embryo development (6, 9). Our analysis shows that the SI-stimulated caspase-like activities are distinct from these enzymes, and the LEVDase represents a plant caspase-like activity not identified previously. We found no evidence implicating activation of caspase-8 or -9 (initiator-type) in SI. Because no reports of this class of caspase exist in plants, this result suggests a possible difference between PCD in animals and plants.
SI Results in Intracellular Acidification.
The SI-induced caspase-like activities exhibit maximum substrate cleavage at physiologically very low pH values, suggesting that the intracellular pH may alter after SI induction. This finding is in contrast to most plant caspase-like activities identified to date, although there are exceptions (13). Cytosolic acidification is a common early apoptotic event (26, 27), although its exact role remains debated (36). Measurement of pollen tube cytosolic [pH]i revealed that it is dramatically altered after SI induction. The extent of the acidification fits the pH optima for the SI-induced caspase activities almost exactly. In plants, vacuolar rupture is a key feature in PCD during xylem formation (37, 38); moreover, PCD in tobacco involving vacuolar processing enzyme, a YVADase (10), presumably involves vacuolar leakage, which would result in a pH drop. However, PCD-associated cytosolic acidification has not been reported in plants.
In summary, our data provide significant advances in our understanding of mechanisms involved in SI in Papaver and also contribute to knowledge about caspase-like enzymes in plant cells. Because plants have no caspase homologs, the nature of their caspase-like proteases is of key importance to an understanding of PCD. Visualization of the SI-induced DEVDase in the cytosol and nuclei, together with demonstration of acidification of the cytosol, provides important evidence for parallels between animal and plant caspase-like protease-mediated events.
Methods
Pollen Tube Growth and SI Induction.
Pollen of P. rhoeas was grown in vitro in liquid germination medium (GM), covering a layer of solidified GM (22) for 1 h before treatments. SI was induced by adding recombinant S proteins (final concentration 10 μg ml−1) to incompatible S3S8 pollen growing in vitro (22). Expression and purification of the recombinant pistil S proteins were as described (22, 39). For each SI-induced sample, a noninduced control was prepared by adding GM. Pollen tubes were grown and harvested 5 h after SI induction, except for temporal studies, where pollen was harvested at various time intervals after SI induction.
Pollen Protein Extraction.
Caspase Extraction (CE) buffer [50 mM sodium acetate/10 mM l-cysteine/10% (vol/vol) glycerol/0.1% wt/vol CHAPS], pH 6.0, was added to pollen. Samples were snap-frozen in liquid N2; thawing samples were homogenized, sonicated, and vortexed and then incubated on ice for 30 min. Samples were centrifuged (16,000 × g, 20 min, 4°C). The protein concentration of the supernatant was determined (Bio-Rad, Hercules, CA).
Caspase Activity Assays.
Caspase-like activities in pollen extracts were measured by using AMC substrates: Ac-DEVD-AMC, Ac-VEID-AMC, Ac-LEVD-AMC, Ac-YVAD-AMC, Ac-IETD-AMC, Ac-LEHD-AMC (EMD Biosciences, San Diego, CA; BIOMOL, Plymouth Meeting, PA), or the MCA substrate Boc-GRR-MCA (Peptide Institute, Inc., Osaka, Japan). Assays contained 10 μg of pollen protein extract and 50 μM fluorogenic substrate. Unless stated otherwise, assays were performed in optimized caspase assay buffer (CE buffer adjusted to pH 5.0). Release of fluorophore by cleavage was measured (excitation 380 nm, emission 460 nm) by using a FLUOstar OPTIMA reader (BMG Labtechnologies, Offenburg, Germany) at 27°C for 5 h. Relative fluorescence unit readings for control samples were subtracted from SI-induced readings. All assays were performed on three independent samples, each measured in triplicate. P values were calculated by using a two-way ANOVA.
To test the effect of caspase inhibitors on caspase substrate cleavage, 50 μM peptide inhibitor, Ac-YVAD-CHO, Ac-VAD-CHO, Ac-DEVD-CHO, Ac-VEID-CHO, Ac-LEVD-CHO, Ac-IETD-CHO, Ac-LEHD-CHO, or Ac-IEPD-CHO (EMD Biosciences and BIOMOL) was added to the caspase assays. To test the effect of cations on caspase-like activities, ZnCl2, CaCl2, MgCl2, NaCl, and KCl were added to the assay reaction mixtures. Sensitivity to protease inhibitors was tested in a similar manner. For determination of optimal pH, 50 mM citrate/phosphate buffers were used. DNA fragmentation was assessed with TUNEL assays according to Thomas and Franklin-Tong (17) by using pretreatment with tetrapeptide inhibitor Ac-DEVD-CHO, Ac-VEID-CHO, or Ac-LEVD-CHO added 1 h before SI induction; for each, 100 pollen tubes were scored in three independent experiments.
Live-Cell Caspase Imaging with NucView 488.
The NucView 488 caspase-3 assay Kit (Biotium, Inc., Hayward, CA) was used to detect DEVDase activity in live pollen tubes. The specificity of the DEVDase activity was checked by pretreating hydrated pollen for 1 h with 100 μM Ac-DEVD-CHO or Ac-YVAD-CHO before SI induction; 2.5 h after SI induction an additional 100 μM inhibitor was added. DMSO was a negative control. One hour before observations were made, the NucView 488 probe (3 μM) was added. Five hours after SI induction, pollen tubes were transferred to 0.05% (wt/vol) poly-l-lysine-coated microscope slides and fixed (50 mM Pipes, pH 6.7/0.5 mM MgCl2/1 mM EGTA/3% formaldehyde), washed with PBS, and mounted with Vectashield (Vector Laboratories, Peterborough, U.K.). For temporal analysis with NucView 488, SI induction times were reduced.
Images were acquired with a Leica DM IRE2 inverted confocal microscope, ×63 oil immersion lens, N.A. 1.4 (Leica Microsystems, Wetzlar, Germany). Nuclear DNA was localized by using DAPI (405-nm blue diode laser; 440–470 BP filter). NucView 488 was imaged with a 488-nm argon laser, 510–550 band pass filter. For each treatment, three independent samples were analyzed; 25 pollen tubes were scored for NucView 488 nuclear staining using identical detection settings. Staining was quantified with ImageJ software (National Institutes of Health, Bethesda, MD). Three categories were obtained by calculating the ratio of fluorescence intensity of nuclei to cytoplasm: ≤1.5, no DEVDase (−); >1.5 ≤2.0, low DEVDase (±); >2.0, high DEVDase (+).
Live-Cell Caspase Imaging with CR(DEVD)2.
SI-induced pollen tubes were incubated with CR(DEVD)2 (CV-Caspase 3&7 detection kit, BIOMOL). After 30 min, pollen tubes were transferred to 0.01% poly-l-lysine-coated slides, excess probe was washed off, and pollen tubes were imaged with confocal microscopy to detect liberated CR fluorescence (Leica; He/Ne laser 594 nm line, 620- to 660-nm band pass filter).
[pH]i Measurements.
[pH]i was imaged in living pollen tubes with the acetoxymethyl ester of the pH-sensitive fluorophore BCECF, using a Leica DM IRE2 confocal microscope. SI-induced pollen tubes were loaded with 2 μM BCECF for 20 min 1–3 h after SI induction and imaged immediately with sequential excitation at 458 nm and 488 nm and emission at 510–550 nm. Untreated pollen tubes were analyzed for comparison. Ratios were calculated, and calibration was carried out by making an in vitro calibration curve according to Feijo et al. (40).
Supplementary Material
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council (to the V.E.F.-T. laboratory).
Abbreviations
- AMC
7-amino-4-trifluoromethyl coumarin
- BCECF
2′-7′-bis(carboxyethyl)-5(6)-carboxyfluorescein
- CR
cresyl violet
- GC
generative cell
- MCA
4-methyl-coumaryl-7-amide
- PCD
programmed cell death
- [pH]i
intracellular cytosolic pH
- SI
self-incompatibility
- VN
vegetative nucleus.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705826104/DC1.
References
- 1.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 2.Sanmartin M, Jaroszewski L, Raikhel NV, Rojo E. Plant Physiol. 2005;137:841–847. doi: 10.1104/pp.104.058552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uren GA, O'Rourke K, Pisabarro TM, Seshgari S, Koonin VE, Dixit VM. Mol Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 4.Woltering EJ. Trends Plants Sci. 2004;9:469–472. doi: 10.1016/j.tplants.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herian M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, Frohlich KU. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- 6.Bozhkov PV, Filonova LH, Suarez MF, Helmersson A, Smertenko AP, Zhivotovsky B, von Arnold S. Cell Death Differ. 2004;11:175–182. doi: 10.1038/sj.cdd.4401330. [DOI] [PubMed] [Google Scholar]
- 7.Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inze D, Van Breusegem F. J Biol Chem. 2004;279:45329–45336. doi: 10.1074/jbc.M406329200. [DOI] [PubMed] [Google Scholar]
- 8.van Doorn WG, Woltering EJ. Trends Plants Sci. 2005;10:117–122. doi: 10.1016/j.tplants.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Boren M, Hoglund A-S, Bozhkov P, Jansson C. J Exp Bot. 2006;57:3747–3753. doi: 10.1093/jxb/erl136. [DOI] [PubMed] [Google Scholar]
- 10.Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I. Science. 2004;305:855–858. doi: 10.1126/science.1099859. [DOI] [PubMed] [Google Scholar]
- 11.Rojo E, Martin R, Carter C, Zouhar J, Pan S, Plotnikova J, Jin H, Paneque M, Sanchez-Serrano JJ, Baker B. Curr Biol. 2004;14:1897–1906. doi: 10.1016/j.cub.2004.09.056. [DOI] [PubMed] [Google Scholar]
- 12.Coffeen WC, Wolpert TJ. Plant Cell. 2004;16:857–873. doi: 10.1105/tpc.017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danon A, Rotari VI, Gordon A, Mailhac N, Gallois P. J Biol Chem. 2004;279:779–787. doi: 10.1074/jbc.M304468200. [DOI] [PubMed] [Google Scholar]
- 14.Thomas SG, Huang S, Li S, Staiger CJ, Franklin-Tong VE. J Cell Biol. 2006;174:221–229. doi: 10.1083/jcb.200604011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.del Pozo O, Lam E. Curr Biol. 1998;8:1129–1132. doi: 10.1016/s0960-9822(98)70469-5. [DOI] [PubMed] [Google Scholar]
- 16.Korthout HAAJ, Berecki G, Bruin W, van Duijn B, Wang M. FEBS Lett. 2000;475:139–144. doi: 10.1016/s0014-5793(00)01643-4. [DOI] [PubMed] [Google Scholar]
- 17.Thomas SG, Franklin-Tong VE. Nature. 2004;429:305–309. doi: 10.1038/nature02540. [DOI] [PubMed] [Google Scholar]
- 18.Takayama S, Isogai A. Annu Rev Plant Biol. 2005;56:467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- 19.McClure B, Franklin-Tong V. Planta. 2006;224:233–245. doi: 10.1007/s00425-006-0284-2. [DOI] [PubMed] [Google Scholar]
- 20.Foote HCC, Ride JP, Franklin-Tong VE, Walker EA, Lawrence MJ, Franklin FCH. Proc Natl Acad Sci USA. 1994;91:2265–2269. doi: 10.1073/pnas.91.6.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franklin-Tong VE, Ride JP, Read ND, Trewavas AJ, Franklin FCH. Plant J. 1993;4:163–177. [Google Scholar]
- 22.Snowman BN, Kovar DR, Shevchenko G, Franklin-Tong VE, Staiger CJ. Plant Cell. 2002;14:2613–2626. doi: 10.1105/tpc.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Graaf BHJ, Rudd JJ, Wheeler MJ, Perry RM, Bell EM, Osman K, Franklin FCH, Franklin-Tong VE. Nature. 2006;444:490–493. doi: 10.1038/nature05311. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Samaj J, Franklin-Tong VE. Plant Physiol. 2007;145:236–245. doi: 10.1104/pp.107.101741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bozhkov PV, Suarez MF, Filonova LH, Daniel G, Zamyatnin AA, Jr, Rodriguez-Nieto S, Zhivotovsky B, Smertenko A. Proc Natl Acad Sci USA. 2005;102:14463–14468. doi: 10.1073/pnas.0506948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuyama S, Reed JC. Cell Death Differ. 2000;7:1155–1165. doi: 10.1038/sj.cdd.4400779. [DOI] [PubMed] [Google Scholar]
- 27.Matsuyama S, Llopis J, Deveraux QL, Tsien RY, Reed JC. Nat Cell Biol. 2000;2:231–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- 28.Stennicke HR, Salvesen GS. J Biol Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 29.Elbaz M, Avni A, Weil M. Cell Death Differ. 2002;9:726–733. doi: 10.1038/sj.cdd.4401030. [DOI] [PubMed] [Google Scholar]
- 30.Faleiro L, Lazebnik Y. J Cell Biol. 2000;151:951–960. doi: 10.1083/jcb.151.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhivotosky B, Samali A, Gahm A, Orrenius S. Cell Death Differ. 1999;6:644–651. doi: 10.1038/sj.cdd.4400536. [DOI] [PubMed] [Google Scholar]
- 32.Jordan ND, Ride JP, Rudd JJ, Davies EM, Franklin-Tong VE, Franklin FCH. Ann Bot (London) 2000;85:197–202. [Google Scholar]
- 33.Tyas L, Brophy VA, Pope A, Rivett AJ, Tavaré JM. EMBO Rep. 2000;1:266–270. doi: 10.1093/embo-reports/kvd050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vacca RA, Valenti D, Bobba A, Merafina RS, Passarella S, Marra E. Plant Physiol. 2006;141:208–219. doi: 10.1104/pp.106.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakaune S, Yamada K, Kondo M, Kato T, Tabata S, Nishimura M, Hara-Nishimura I. Plant Cell. 2005;17:876–887. doi: 10.1105/tpc.104.026872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagadic-Gossmann D, Huc L, Lecureur V. Cell Death Differ. 2004;11:953–961. doi: 10.1038/sj.cdd.4401466. [DOI] [PubMed] [Google Scholar]
- 37.Kuriyama H, Fukuda H. Curr Opin Plant Biol. 2004;5:568–573. doi: 10.1016/s1369-5266(02)00305-9. [DOI] [PubMed] [Google Scholar]
- 38.Obara K, Kuriyama H, Fukuda H. Plant Physiol. 2001;125:615–626. doi: 10.1104/pp.125.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foote HCC, Ride JP, Franklintong VE, Walker EA, Lawrence MJ, Franklin FCH. Proc Natl Acad Sci USA. 1994;91:2265–2269. doi: 10.1073/pnas.91.6.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feijo JA, Sainhas J, Hackett GR, Kunkel JG, Hepler PK. J Cell Biol. 1999;144:483–496. doi: 10.1083/jcb.144.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.