Abstract
Haemophilus ducreyi is a Gram-negative bacterium that causes chancroid, a sexually transmitted genital ulcer disease. Different lipooligosaccharide (LOS) structures have been identified from H. ducreyi strain 35000, including those sialylated glycoforms. Surface LOS of H. ducreyi is considered an important virulence factor that is involved in ulcer formation, cell adhesion, and invasion of host tissue. Gene Hd0686 of H. ducreyi, designated lst (for lipooligosaccharide sialyltransferase), was identified to encode an α2,3-sialyltransferase that is important for the formation of sialylated LOS. Here we show that Hd0053 of H. ducreyi genomic strain 35000HP, the third member of the glycosyltransferase family 80 (GT80), also encodes an α2,3-sialyltransferase that may be important for LOS sialylation.
Keywords: bacterial sialyltransferase, glycosyltransferase, Haemophilus ducreyi, GT80, lipooligosaccharides, sialyltransferase
Introduction
Haemophilus ducreyi is a Gram-negative human mucosal pathogen that causes chancroid, a highly contagious sexually transmitted genital ulcer disease [1]. Chancroid is common in developing countries. Although less common in the United States, chancroid has been associated with increased risk for developing other sexually transmitted diseases including human immunodeficiency virus (HIV) [2]. Sialylated LOS structures have been identified from H. ducreyi strains and may be expressed by the bacteria to mimic host cell surface glycans to evade the host's immune system as other Gram-negative mucosal pathogens, such as Haemophilus influenzae, Neisseria meningitidis, and Neisseria gonorrhoeae [3]. The lst gene in H. ducreyi (Hd0686) has been proven to be a lipooligosaccharide (LOS) sialyltransferase gene by complementation studies [4]. It shares homology to cpsK gene, a capsular polysaccharide sialyltransferase gene, in GBS strains [5] and Hi0871 gene of H. influenzae, which encodes a hypothetic lipooligosaccharide sialyltransferase [6]. H. ducreyi is suggested to use a sialic acid scavenging mechanism for LOS sialylation which is similar to that described for H. influenzae and Haemophilus somnus [7-10]. This precursor scavenging pathway involves the uptake of host sialic acid as the precursor. Production of activated sugar nucleotide donor CMP-sialic acid and subsequent transfer of sialic acid to appropriate membrane acceptors are then accomplished by CMP-sialic acid synthetase(s) and sialyltransferase(s) produced by the bacteria [8]. While the LOS from the H. ducreyi neuA (a CMP-N-acetylneuraminic acid synthetase gene) mutant lacked detectable sialic acid, the LOS from the lst mutant still contained some sialic acid [4]. This indicates that another sialyltransferase may exist that contributes to the sialylation of LOS in H. ducreyi.
BLAST search using the amino acid sequence of a Photobacterium damsela α2,6-sialyltransferase (Pd2,6ST or sialyltransferase 0160) (GenBank accession number: BAA25316) [11,12] identified two homologous proteins encoded by gene Pm0188 from Pasteurella multocida genomic strain Pm70 and gene Hd0053 from the H. ducreyi genomic strain 35000HP, respectively. These three proteins do not share sequence homology with any other reported bacterial or mammalian glycosyltransferases. They have now been classified into a new glycosyltransferase family GT80 on the Carbohydrate-Active enzymes (CAZy) database (http://afmb.cnrs-mrs.fr/CAZY/) [13]. We have shown previously that Pm0188 encodes a multifunctional sialyltransferase having α2,3-sialyltransferase, α2,6-sialyltransferase, α2,3-sialidase, and α2,3-trans-sialidase functions [14]. Here, we report the cloning and biochemical characterization of the Hd0053 gene product. The recombinant enzyme was expressed as a His-tagged fusion protein and purified to homogeneity using a Ni-NTA affinity column. NMR studies of the product produced by the reaction catalyzed by the enzyme confirm that the Hd0053 gene encodes an α2,3-sialyltransferase. This is the second α2,3-sialyltransferase identified from H. ducreyi.
Materials and methods
Bacterial strains, plasmids, and materials.
E. coli electrocompetent DH5α and chemically competent BL21 (DE3) cells were from Invitrogen (Carlsbad, CA). Genomic DNA of H. ducreyi 35000HP was from American Type Culture Collection (ATCC, Manassas, VA) (ATCC#700724D). Vector plasmids pET15b and pET22b(+) were purchased from Novagen (EMD Biosciences, Inc. Madison,WI). Ni2+–NTA agarose (nickel–nitrilotriacetic acid–agarose), QIAprep spin miniprep kit, and QIAEX II gel extraction kit were from Qiagen (Valencia, CA). Herculase enhanced DNA polymerase was from Stratagene (La Jolla, CA). T4 DNA ligase, 1 kb DNA ladder, and BamHI restriction enzyme were obtained from Promega (Madison, WI). NdeI restriction enzyme was from New England Biolabs, Inc. (Beverly, MA). Precision Plus Protein Standards, and BioGel P-2 fine resin were from Bio-Rad (Hercules, CA). Bicinchoninic acid (BCA) protein assay kit was from Pierce Biotechnology, Inc. (Rockford, IL). CTP, D-N-acetylmannosamine (ManNAc), and pyruvate were purchased from Sigma (St. Louis, MO). CMP-Neu5Ac was synthesized enzymatically from ManNAc, pyruvate, and CTP by a one-pot two-enzyme system using a recombinant sialic acid aldolase cloned from E. coli K12 and a recombinant N. meningitidis CMP-sialic acid synthetase as described previously [15]. 4-Methylumbelliferyl β-D-lactoside (LacMU) and 3-azidopropyl β-D-lactoside (LacβProN3) were synthesized from lactose through a hepta-O-acetyllactosyl trichloroacetimidate intermediate as reported [14].
Cloning.
Hd0053 was cloned as an N-His6-tagged or a C-His6-tagged fusion protein using genomic DNA of H. ducreyi strain 35000HP as the template for polymerase chain reactions (PCR). The primers used to clone the N-His-tagged protein in pET15b vector were: forward primer 5'GATCCATATGCTGATTCAACAAAATCTTG (NdeI restriction site is underlined) and reverse primer 5' CGCGGATCCTTAATTATGTATTGTACACATAAATGC 3' (BamHI restriction site is underlined). To clone the C-His-tagged protein in pET22b(+) vector, the same forward primer was used, and the reverse primer used was 5' CGCGGATCCTTAGTGATGATGATGATGATGATTATGTATTGTACACATAAATGC 3' (BamHI restriction site is underlined, sequence that encodes the hexahistidine tag is in italics.). PCR reactions for amplifying the target gene were performed in a 50 μL reaction mixture containing genomic DNA (1 μg), forward and reverse primers (1 μM each) , 10 × Herculase buffer (5 μL), dNTP mixture (1 mM), and 5 units (1 μL) of Herculase enhanced DNA polymerase. The reaction mixture was subjected to 35 cycles of amplification with an annealing temperature of 52 °C. The resulting PCR product was purified and digested with NdeI and BamHI restriction enzymes. The purified and digested PCR product was ligated with predigested pET15b or pET22b(+) vector and transformed into electrocompetent E. coli DH5αcells. Selected clones were grown for minipreps and characterization by restriction mapping and DNA sequencing performed by Davis Sequencing Facility in the University of California-Davis.
Expression.
Positive plasmid was selected and transformed into BL21 (DE3) chemically competent cells. The plasmid-bearing E. coli strain was cultured in LB rich medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) supplemented with ampicillin (100 μg/mL). Overexpression of the target protein was achieved by inducing the E. coli culture with 0.1 mM of isopropyl-1-thio-β-D-galactopyranoside (IPTG) when the OD 600 nm of the culture reaches 0.8−1.0 and incubating at 15 °C for 48 h with vigorous shaking at 250 rpm in a C25KC incubator shaker (New Brunswick Scientific, Edison, NJ).
Purification.
His6-tagged target proteins were purified from cell lysate. To obtain the cell lysate, cell pellet harvested by centrifugation at 4,000 rpm for 2 hrs was resuspended in lysis buffer (pH 8.0, 100 mM Tris–HCl containing 0.1% Triton X-100) (20 mL per liter cell culture). Lysozyme (50 μg/mL) and DNaseI (3 μg/mL) were then added and the mixture was incubated at 37 °C for 50 min with vigorous shaking. Cell lysate was obtained by centrifugation at 11,000 rpm for 30 min as the supernatant. Purification of His-tagged proteins from the lysate was achieved using an AKTA FPLC system (GE Healthcare) equipped with a HisTrap™ FF 5 mL column. The column was pre-equilibrated with 10 column volumes of binding buffer (5 mM imidazole, 0.5 M NaCl, 50 mM Tris–HCl, pH 7.5) before the lysate was loaded. Followed by washing with 8 column volumes of binding buffer, 10 column volumes of washing buffer (50 mM imidazole, 0.5 M NaCl, 50 mM Tris–HCl, pH 7.5), the protein was eluted with a linear gradient of elute buffer containing 50−250 mM imidazole in Tris-HCl buffer (50 mM, pH 7.5, 0.5 M NaCl). The fractions containing the purified enzymes were collected and stored at 4 °C.
pH Profile by HPLC.
Typical enzymatic assays were performed in a total volume of 20 μL in a buffer (200 mM) with pH varying from 5.0−11.0 containing CMP-Neu5Ac (2 mM), LacMU (1 mM), and the recombinant enzyme (0.3 μg). The buffers used were: MES, pH 5.0−6.0; Hepes, pH 7.0; Tris–HCl, pH 7.5−9.0; and CAPS, pH 10−11. Reactions were allowed to proceed for 60 min at 37 °C and quenched by adding ice-cold 12% acetonitrile (780 μL) to make 40-fold dilution. The samples were then kept on ice until aliquots of 10 μL were injected and analyzed by a Shimadzu LC-2010A system equipped with a membrane on-line degasser, a temperature control unit and a fluorescence detector. A reverse phase Premier C18 column (250 × 4.6 mm I.D., 5 μm particle size, Shimadzu) protected with a C18 guard column cartridge was used. The mobile phase was 12% acetonitrile. The fluorescent compounds LacMU and Neu5Acα2,3LacMU were detected by excitation at 325 nm and emission at 372 nm [16]. All assays were carried out in duplicate.
Effects of metal ions, EDTA, and dithiothreitol (DTT).
EDTA (5 mM), different concentrations (5 mM, 10 mM, and 20 mM) of MgCl2 or MnCl2, and various concentrations of DTT (0 mM, 0.2 mM, 1 mM, and 5 mM) were used in a Tris–HCl buffer (pH 8.5, 100 mM) to analyze their effects on the α2,3-sialyltransferase activity of Hd0053 (0.35 μg). Reaction without EDTA, DTT, and metal ions was used as a control.
Kinetics by HPLC assays.
The enzymatic assays were carried out in a total volume of 20 μL in a Tris-HCl buffer (100 mM, pH 8.5) containing CMP-Neu5Ac, LacMU and the recombinant protein (0.3 μg). Reactions were allowed to proceed for 60 min at 37 °C. Apparent kinetic parameters were obtained by varying the CMP-Neu5Ac concentration from 0.1−4.0 mM (0.1mM, 0.25 mM, 0.4 mM, 1 mM, 2 mM, and 4 mM) and a fixed concentration of LacMU (1 mM), or a fixed concentration of CMP-Neu5Ac (1 mM) and varied concentrations of LacMU (0.1 mM, 0.25 mM, 0.4 mM, 1 mM, 2 mM, and 4 mM). The double reciprocal Lineweaver-Burk plots were obtained from the average values of duplicate or triplet assay results. One unit of enzyme is defined as the amount of enzyme that synthesizes the formation of 1 μmole of Neu5Acα2,3LacMU per minute at 37 °C under the assay conditions.
Enzymatic synthesis of Neu5Acα2,3LacβProN3.
The synthesis was carried out in a total volume of 10 mL in Tris-HCl buffer (100 mM, pH 8.5) containing LacβProN3 (27 mg, 0.063 mmol), CMP-Neu5Ac (50 mg, 0.076 mmol), and Hd0053 (25 μg). The reaction was allowed to proceed at 37 °C for 2 hrs and further reacted at room temperature for overnight when TLC analysis (developing solvent used was EtOAc : MeOH : H2O : HOAc = 4 : 2 : 1 : 0.1) indicated the completion of the reaction. The reaction was stopped by adding an equal volume of ice-cold ethanol. The mixture was mixed well and kept on ice for 30 min. After the precipitated protein was removed by centrifuging at 5000 × g for 30 min, the supernatant was concentrated and purified by Bio-Gel P2 size exclusion chromatography and lyophilized to give Neu5Acα2,3LacβProN3 (38 mg, 82% yield) as white powder. 1H-NMR, 1H-1 H COSY, and 1H-13C HSQC experiments were carried out at 26 °C in D2O on a Bruker DRX-600 and 13C-NMR experiment was carried out at Varian mercuryplus 300 spectrometer. The proton chemical shifts were referenced to the HOD signal at 4.79 ppm and the 13C chemical shifts were referenced to the methyl resonance of the internal deuterated acetone at 30.89 ppm.
Results
Expression and purification of Hd0053 protein
Hd0053 from H. ducreyi strain 35000HP was cloned as both an N-His6-tagged and a C-His6-tagged recombinant proteins using pET15b and pET22b(+) vectors, respectively. The DNA sequence of the cloned gene matches to that reported in GenBank (Accession number AE017143). The optimized expression condition was incubating at 15 °C for 48 h with vigorous shaking (250 rpm) after induction with 0.1 mM of IPTG. Under the same expression conditions, the expression level of N-His6-tagged form is higher than that of the C-His6-tagged one. Therefore, only the N-His6-tagged Hd0053 is characterized in detail. The SDS-PAGE analysis of the protein expression (Fig. 1.) indicates that Hd0053 was expressed in a large amount which consists of about 70% of the total protein extracts of the E. coli host cells. The solubility of the recombinant protein, however, was quite low. Only a small portion of the target protein was observed in the lysate, which is the soluble portion of the protein. Ni-Column purification using the AKTA FPLC system provided purified protein showing a molecular weight of about 42 kD in the SDS-PAGE. This molecular weight is close to that (48.7 kD) calculated for the Hd0053 protein.
Fig. 1.
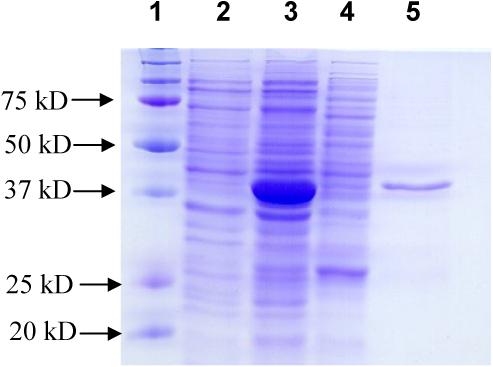
SDS-PAGE analysis for Hd0053 protein expression and purification. Lanes: 1, protein standards; 2, whole cell extraction before induction; 3, whole cell extraction after induction; 4, lysate after induction; 5, Ni-column purified Hd0053 protein.
pH Profile of Hd0053
The pH profile of the α2,3-sialyltransferase activity of Hd0053 (Fig. 2A) indicates that the enzyme catalyzes the transfer of Neu5Ac from CMP-Neu5Ac to an acceptor such as LacMU under basic condition (pH > 7.0) with a optimal activity at pH 8.0. More than 90% of the activity was observed at pH 9.0. The enzyme activity declines quickly when pH is below 7.5 or above 9.0. The activity drops to about 16% and 18% when the pH is at 5.0 and 11.0, respectively.
Fig. 2.
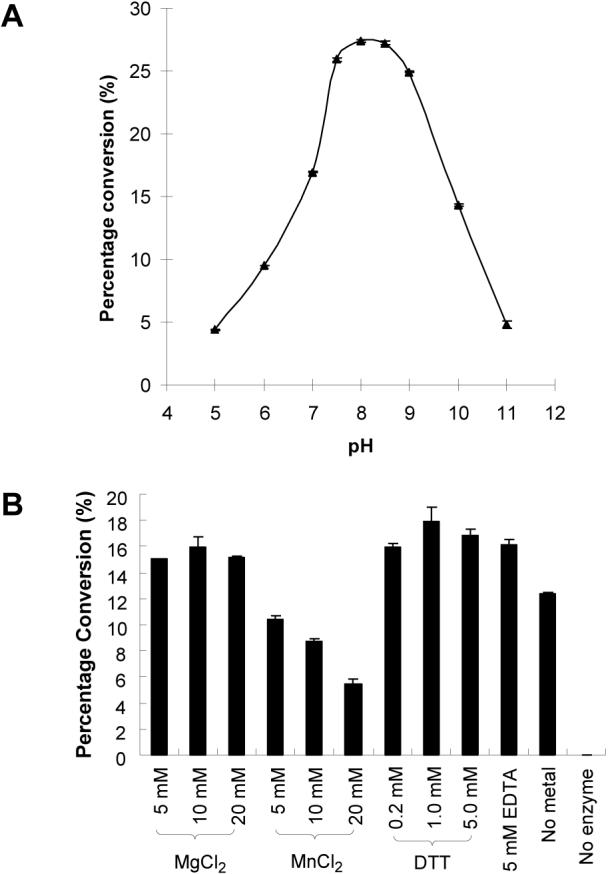
The pH profile and the effects of metal ions, EDTA, and DTT on the α2,3-sialyltransferase activity of Hd0053 by quantitative HPLC analysis. (A) The pH profile. Activity was measured at indicated pH at 37 °C for 60 min. Buffers (200 mM) used were: MES, pH 5.0−6.0; Hepes, pH 7.0; Tris–HCl, pH 7.5−9.0; and CAPS, pH 10−11. (B) Effects of metal ions, EDTA, and DTT. The activity was determined in Tris-HCl buffer (100 mM, pH 8.5) at 37 °C for 60 min.
Effect of metal ions, EDTA, and dithiothreitol (DTT) on the activity of Hd0053
The effects of various metal ions, the chelating agent EDTA, and DTT on the enzyme activity of Hd0053 were examined at pH 8.5. As shown in Fig. 2B, a divalent metal ion is not required for the α2,3-sialyltransferase activity of the enzyme, as 5 mM of EDTA does not affect the enzyme activity. The addition of Mg2+ up to 20 mM does not affect the activity of the enzyme. Increasing the concentration of Mn2+ in the reaction mixture, however, decreases the activity of the enzyme. The detrimental effect of the Mn2+ is similar to that was shown for the α2,3-sialyltransferase activity of PmST1, but is not as dramatic [14]. The metal effect is different from that reported for the N. meningitidis (MC58 and 406Y) Lst, for which the sialyltransferase activity can be stimulated by Mg2+ (2-fold) or Mn2+ (3-fold) [17].
There are four non-conserved cysteine residues in the Hd0053 protein sequence. The effect of DTT on the α2,3-sialyltransferase activity of the enzyme was studied. Addition of DTT up to 5 mM does not significantly increase the activity of Hd0053 (Fig. 2B), indicating disulfide formation is not required for the sialyltransferase activity of the Hd0053.
Kinetics.
The apparent KM values obtained for the α2,3-sialyltransferase activity of Hd0053 are 0.05 mM and 5.3 mM for CMP-Neu5Ac and LacMU, respectively (Table 1). This KM value of CMP-Neu5Ac (0.05 mM) is comparable to that for recombinant N. meningitidis α2,3-sialyltransferase (20 μM) [18], rat liver Galβ1,4GlcNAc α2,6-sialyltransferase (50 μM), and Galβ1,4(3)GlcNAc α2,3-sialyltransferase (70 μM) [19]. It is one magnitude less than that of PmST1 (0.44 mM) [14] and Cst II (0.46 mM) [20]. The KM value of LacMU (5.3 mM) for Hd0053 is three fold to that for the PmST1 (1.4 mM), indicating a weaker binding of the LacMU to Hd0053 than to PmST1 [14]. The kcat/KM for LacMU is 0.13 s−1 mM−1 (7.8 min−1 mM−1) for the Hd0053, which is about three fold less than that (22 min−1 mM−1) obtained for APTS (8-aminopyrene-1,3,6-trisulfonic acid)-labeled lactose for the N. meningitidis α2,3-sialyltransferase [18]. It is 254 fold less than that of the α2,3-sialyltransferase activity of the PmST1 (33 s−1 mM−1) [14].
Table 1.
Apparent kinetic parameters for the α2,3-sialyltransferase activity of Hd0053.a
| Substrates | CMP-Neu5Ac | LacMU |
|---|---|---|
| KM (mM) | 0.05 ± 0.008 | 5.3 ± 0.46 |
| Vmax (mM s−1) | (3.0 ± 0.05) × 10−5 | (2.0 ± 0.08) × 10−4 |
| kcat (s−1) | 0.10 ± 0.002 | 0.67 ± 0.03 |
| kcat/KM (s−1 mM−1) | 2.0 | 0.13 |
Assays were performed in triplicate in Tris-HCl buffer (100 mM, pH 8.5) with either varied concentrations of CMP-Neu5Ac (0.1, 0.25, 0.4, 1, 2, 4 mM) and a fixed concentration of LacMU (1 mM) or a fixed concentration of CMP-Neu5Ac (1 mM) and varied concentrations (0.1, 0.25, 0.4, 1, 2, and 4 mM) of LacMU. Hd0053 was used at the same concentration (0.3 μM) for these assays.
Enzymatic synthesis and characterization of sialylation product
Using LacβProN3 as the acceptor and CMP-Neu5Ac as the donor, the synthesis of sialylated LacβProN3 was achieved in high yield (82%) at pH 8.5. The purified sialylated product was characterized by NMR spectrometry. As listed in Table 2, the NMR data match well to those reported for α2,3-sialosides [14,21]. For example, the chemical shift of the proton at C-3 of Gal in the sialylated product (4.09 ppm) is 0.39−0.46 ppm downfield to that in Neu5Acα2,6LacβProN3 [12] and LacβProN3 [14]. The difference of the chemical shifts of two protons at C-6 of Gal in the sialylated product (0.05 ppm) is similar to that in LacβProN3 [14], but is much smaller than that in Neu5Acα2,6LacβProN (0.35 ppm) [12]. In the 13C NMR spectrum, the chemical shifts of C-3 and C-2 on the Gal of the sialylated product are 2.7 ppm downfield and 1.5 ppm upfield, respectively, compared to those in LacβN3 (72.9 ppm). All other 13C chemical shifts for the LacβProN3 moiety in the sialylated product are very similar (within 0.5 ppm difference) to those in LacβProN3 [14]. These data strongly confirm that the sialylation product catalyzed by Hd0053 is α2,3-linked sialoside Neu5Acα2,3LacβProN3.
Table 2.
Chemical shifts (ppm) and assignments of Neu5Acα2,3LacβProN3, the sialylation product catalyzed by H. ducreyi α2,3-sialyltransferase Hd0053.a
| Sugar | Position | H | C |
|---|---|---|---|
| Glc | 1 | 4.46 | 102.3 |
| 2 | 3.29 | 72.9 | |
| 3 | 3.63 | 74.5 | |
| 4 | 3.64 | 78.3 | |
| 5 | 3.58 | 74.9 | |
| 6 | 3.96 | 60.2 | |
| 6' | 3.80 | ||
| Propyl azide | -CH2-CH2-CH2-N3 | 3.99 | 67.5 |
| 3.76 | |||
| -CH2-CH2-CH2-N3 | 1.89 | 28.4 | |
| -CH2-CH2-CH2-N3 | 3.43 | 48.0 | |
| Gal | 1 | 4.50 | 102.8 |
| 2 | 3.55 | 69.5 | |
| 3 | 4.09 | 75.6 | |
| 4 | 3.94 | 67.6 | |
| 5 | 3.69 | 75.3 | |
| 6 | 3.74 | 61.2 | |
| 6' | 3.69 | ||
| Neu5Ac | 1 | 174.0 | |
| 2 | 99.9 | ||
| 3ax | 1.77 | 39.8 | |
| 3eq | 2.73 | ||
| 4 | 3.67 | 68.5 | |
| 5 | 3.84 | 51.8 | |
| 6 | 3.62 | 73.0 | |
| 7 | 3.57 | 68.2 | |
| 8 | 3.88 | 71.9 | |
| 9 | 3.86 | 62.7 | |
| 9' | 3.66 | ||
| N-CO-CH3 | 2.01 | 22.2 | |
| N-CO-CH3 | 175.1 |
NMR spectra were recorded at 299 K in D2O at neutral pH. Chemical shifts (ppm) are given relative to HOD (4.79 ppm) for 1H NMR or the methyl resonance of the internal deuterated acetone (30.89 ppm) for 13C NMR. The peaks were assigned using 1H-1H COSY and 1H-13C HSQC with reference to [21].
Discussion
Sialic acids are commonly found as terminal carbohydrate residues on cell surface glycoconjugates (glycoproteins and glycolipids) of higher animals. As the terminal carbohydrate residue, sialic acid is one of the first molecules encountered in cellular interactions and has been found to play important roles in cellular recognition and communication [22,23]. Cell surface sialic acids have also been found in relatively few microorganisms, mainly pathogenic bacteria, and their presence is often associated with virulence [22-24]. Sialic acids on LOS of several mucosal pathogens, including N. meningitidis, N. gonorrhoeae, H. ducreyi and H. influenzae strains are considered important LOS components for molecular mimicry to allow them to evade the host's immune system or as modulators of microbial interactions with host cells [7,8,23,25,26]. Some of these pathogenic bacteria present their LOS structure in a phase-variable manner and the degree of sialylation is related to growth conditions of the bacteria [26-28].
Cloning, expression, and biochemical characterization of the Hd0053 gene product shown here confirmed that the Hd0053 gene sequence encodes an active α2,3-sialyltransferase. It is the second sialyltransferase that has been identified in Haemophilus ducreyi 35000HP (the first one is Lst) [4]. Considering the existence of four sialyltransferase genes (siaA, lic3A, lic3B, and lsgB) described in H. influenzae [6,26,29] and three putative sialyltransferase genes predicted in P. multocida [30], the presence of multiple sialyltransferase genes seems to be a common phenomenon for the Haemophilus-Actinobacillus-Pasteurella (HAP) group bacteria. The presence of multiple sialyltransferases in these pathogenic bacteria indicates that bacterial sialylation is a very complex process.
Hd0053 shares 32% identity and 52% similarity to the amino acid sequence of PmST1. Compared to the amino acid sequence of Pd2,6ST, the sequence of Hd0053 has 26% identify and 44% similarity. PmST1, Pd2,6ST, and Hd0053 represent a new family of sialyltransferases since they lack sequence similarity to any other reported sialyltransferases, either from bacterial or mammalian sources. They have now been classified into glycosyltransferase family GT80 on the CAZy database (http://afmb.cnrs-mrs.fr/CAZY/) [13]. Despite the sequence homology shared among these three proteins, they have different activities. While both Pd2,6ST and Hd2,3ST have a single but different ST activity, PmST1 is a multifunctional enzyme for which four different functions have been identified, including an α2,3-sialyltransferase, an α2,6-sialyltransferase, an α2,3-sialidase, and an α2,3-trans-sialidase activities. These three enzymes, thus, provide an excellent model system for structure-function relationship studies of sialyltransferases.
Acknowledgments
This work was supported by NIH R01GM076360 and the start-up funds from the Regents of the University of California.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morse SA. Chancroid and Haemophilus ducreyi. Clin. Microbiol. Rev. 1989;2:137–157. doi: 10.1128/cmr.2.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annan NT, Lewis DA. Treatment of chancroid in resource-poor countries. Expert. Rev. Anti. Infect. Ther. 2005;3:295–306. doi: 10.1586/14787210.3.2.295. [DOI] [PubMed] [Google Scholar]
- 3.Melaugh W, Campagnari AA, Gibson BW. The lipooligosaccharides of Haemophilus ducreyi are highly sialylated. J. Bacteriol. 1996;178:564–570. doi: 10.1128/jb.178.2.564-570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bozue JA, Tullius MV, Wang J, Gibson BW, Munson RS., Jr. Haemophilus ducreyi produces a novel sialyltransferase. Identification of the sialyltransferase gene and construction of mutants deficient in the production of the sialic acid-containing glycoform of the lipooligosaccharide. J. Biol. Chem. 1999;274:4106–4114. doi: 10.1074/jbc.274.7.4106. [DOI] [PubMed] [Google Scholar]
- 5.Chaffin DO, McKinnon K, Rubens CE. CpsK of Streptococcus agalactiae exhibits α2,3-sialyltransferase activity in Haemophilus ducreyi. Mol. Microbiol. 2002;45:109–122. doi: 10.1046/j.1365-2958.2002.02988.x. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Samuels NM, Phillips NJ, et al. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J. Biol. Chem. 2002;277:14598–15611. doi: 10.1074/jbc.M110986200. [DOI] [PubMed] [Google Scholar]
- 7.Mandrell RE, McLaughlin R, Aba Kwaik Y, Lesse A, Yamasaki R, Gibson B, Spinola SM, Apicella MA. Lipooligosaccharides (LOS) of some Haemophilus species mimic human glycosphingolipids, and some LOS are sialylated. Infect. Immun. 1992;60:1322–1328. doi: 10.1128/iai.60.4.1322-1328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vimr E, Lichtensteiger C. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 2002;10:254–257. doi: 10.1016/s0966-842x(02)02361-2. [DOI] [PubMed] [Google Scholar]
- 9.Schilling B, Goon S, Samuels NM, Gaucher SP, Leary JA, Bertozzi CR, Gibson BW. Biosynthesis of sialylated lipooligosaccharides in Haemophilus ducreyi is dependent on exogenous sialic acid and not mannosamine. Biochemistry. 2001;40:12666–12677. doi: 10.1021/bi0107849. [DOI] [PubMed] [Google Scholar]
- 10.Inzana TJ, Glindemann G, Cox AD, Wakarchuk W, Howard MD. Incorporation of N-acetylneuraminic acid into Haemophilus somnus lipooligosaccharide (LOS): enhancement of resistance to serum and reduction of LOS antibody binding. Infect. Immun. 2002;70:4870–4879. doi: 10.1128/IAI.70.9.4870-4879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto T, Nakashizuka M, Terada I. Cloning and expression of a marine bacterial beta-galactoside alpha2,6-sialyltransferase gene from Photobacterium damsela JT0160. J. Biochem. (Tokyo) 1998;123:94–100. doi: 10.1093/oxfordjournals.jbchem.a021921. [DOI] [PubMed] [Google Scholar]
- 12.Yu H, Huang S, Chokhawala H, Sun M, Zheng H, Chen X. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: a P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew. Chem. Int. Ed. Engl. 2006;45:3938–3944. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 14.Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. A multifunctional Pasteurella multocida sialyltransferase: a powerful tool for the synthesis of sialoside libraries. J. Am. Chem. Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Yu H, Karpel R, Chen X. Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorg. Med. Chem. 2004;12:6427–6435. doi: 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 16.Kajihara Y, Kamiyama D, Yamamoto N, Sakakibara T, Izumi M, Hashimoto H. Synthesis of 2-[(2-pyridyl)amino]ethyl beta-D-lactosaminide and evaluation of its acceptor ability for sialyltransferase: a comparison with 4-methylumbelliferyl and dansyl beta-D-lactosaminide. Carbohydr. Res. 2004;339:1545–1550. doi: 10.1016/j.carres.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert M, Watson DC, Cunningham AM, Jennings MP, Young NM, Wakarchuk WW. Cloning of the lipooligosaccharide alpha-2,3-sialyltransferase from the bacterial pathogens Neisseria meningitidis and Neisseria gonorrhoeae. J. Biol. Chem. 1996;271:28271–28276. doi: 10.1074/jbc.271.45.28271. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert M, Cunningham AM, Watson DC, Martin A, Richards JC, Wakarchuk WW. Characterization of a recombinant Neisseria meningitidis alpha-2,3-sialyltransferase and its acceptor specificity. Eur. J. Biochem. 1997;249:187–194. doi: 10.1111/j.1432-1033.1997.t01-1-00187.x. [DOI] [PubMed] [Google Scholar]
- 19.Gross HJ, Rose U, Krause JM, Paulson JC, Schmid K, Feeney RE, Brossmer R. Transfer of synthetic sialic acid analogues to N- and O-linked glycoprotein glycans using four different mammalian sialyltransferases. Biochemistry. 1989;28:7386–7392. doi: 10.1021/bi00444a036. [DOI] [PubMed] [Google Scholar]
- 20.Chiu CP, Watts AG, Lairson LL, Gilbert M, Lim D, Wakarchuk WW, Withers SG, Strynadka NC. Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat. Struct. Mol. Biol. 2004;11:163–170. doi: 10.1038/nsmb720. [DOI] [PubMed] [Google Scholar]
- 21.Berman E. Structural and conformational analysis of sialyloligosaccharides using carbon-13 nuclear magnetic resonance spectroscopy. Biochemistry. 1984;23:3754–3759. doi: 10.1021/bi00311a029. [DOI] [PubMed] [Google Scholar]
- 22.Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 23.Schauer R. Achievements and challenges of sialic acid research. Glycoconj. J. 2000;17:485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith H, Parsons NJ, Cole JA. Sialylation of neisserial lipopolysaccharide: a major influence on pathogenicity. Microb. Pathog. 1995;19:365–377. doi: 10.1006/mpat.1995.0071. [DOI] [PubMed] [Google Scholar]
- 25.Mandrell RE, Apicella MA. Lipo-oligosaccharides (LOS) of mucosal pathogens: molecular mimicry and host-modification of LOS. Immunobiology. 1993;187:382–402. doi: 10.1016/S0171-2985(11)80352-9. [DOI] [PubMed] [Google Scholar]
- 26.Bouchet V, Hood DW, Li J, et al. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8898–8903. doi: 10.1073/pnas.1432026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jennings MP, Srikhanta YN, Moxon ER, Kramer M, Poolman JT, Kuipers B, van der Ley P. The genetic basis of the phase variation repertoire of lipopolysaccharide immunotypes in Neisseria meningitidis. Microbiology. 1999;145(Pt 11):3013–3021. doi: 10.1099/00221287-145-11-3013. [DOI] [PubMed] [Google Scholar]
- 28.Burch CL, Danaher RJ, Stein DC. Antigenic variation in Neisseria gonorrhoeae: production of multiple lipooligosaccharides. J. Bacteriol. 1997;179:982–986. doi: 10.1128/jb.179.3.982-986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox KL, Cox AD, Gilbert M, et al. Identification of a bifunctional lipopolysaccharide sialyltransferase in Haemophilus influenzae. Incorporation of disialic acid. J. Biol. Chem. 2006;281:40024–40032. doi: 10.1074/jbc.M602314200. [DOI] [PubMed] [Google Scholar]
- 30.St Michael F, Vinogradov E, Li J, Cox AD. Structural analysis of the lipopolysaccharide from Pasteurella multocida genome strain Pm70 and identification of the putative lipopolysaccharide glycosyltransferases. Glycobiology. 2005;15:323–333. doi: 10.1093/glycob/cwi015. [DOI] [PubMed] [Google Scholar]


