Abstract
Roles for Eph receptor tyrosine kinase and ephrin signaling in vertebrate brain development are well established. Their involvement in the modulation of mammalian synaptic structure and physiology is also emerging. However, less is known of their effects on brain development and their function in adult invertebrate nervous systems. Here, we report on the characterization of Eph receptor and ephrin orthologs in the honeybee, Apis mellifera (Am), and their role in learning and memory. In situ hybridization for mRNA expression showed a uniform distribution of expression of both genes across the developing pupal and adult brain. However, in situ labeling with Fc fusion proteins indicated that the AmEphR and Amephrin proteins were differentially localized to cell body regions in the mushroom bodies and the developing neuropiles of the antennal and optic lobes. In adults, AmEphR protein was localized to regions of synaptic contacts in optic lobes, in the glomeruli of antennal lobes, and in the medial lobe of the mushroom body. The latter two regions are involved in olfactory learning and memory in the honeybee. Injections of EphR-Fc and ephrin-Fc proteins into the brains of adult bees, 1 h before olfactory conditioning of the proboscis extension reflex, sig-nificantly reduced memory 24 h later. Experimental amnesia in the group injected with ephrin-Fc was apparent 1 h post-training. Experimental amnesia was also induced by post-training injections with ephrin-Fc suggesting a role in recall. This is the first demonstration that Eph molecules function to regulate the formation of memory in insects.
Keywords: Eph receptor, ephrin, associative learning, development, insect
INTRODUCTION
Learning and memory formation underlie the ability of animals to adapt behaviorally to changes in their environment. This depends on the brain ability to reorganize its synaptic structure and function. Molecular mechanisms that underlie specific changes are products of gene families encoding intracellular, membrane bound, and scaffolding proteins. Among them are the Eph receptor (EphR) tyrosine kinases, and their membrane attached ephrin ligands (Murai and Pasquale, 2004). Vertebrates express a large family of EphR and ephrin proteins, subgrouped into either class-A or class-B. The EphA-type receptors are primarily activated by ephrins-As, whereas EphB receptors bind ephrin-B ligands. The mode of membrane attachment for the ephrin-A ligands is glycosyl-phosphatidylinositol-anchor (GPI), whereas the ephrin-B ligands attach to the membrane via a transmembrane domain (Wilkinson, 2000; Martinez and Soriano, 2005). EphR and ephrins are known to regulate numerous events such as tissue patterning (Xu et al., 1995), cell migration (Krull et al., 1997), and axon fasciculation (Winslow et al., 1995) and guidance (Drescher et al., 1995) during neural development. Beyond the period of initial axonal innervation, Eph proteins promote the development of functional synapses (Torres et al., 1998; Buchert et al., 1999; Dalva et al., 2000; Ethell et al., 2001). Indeed, EphA and EphB receptor and ephrin proteins continue to be expressed throughout the mature mammalian brain (Liebl et al., 2003), to regulate structural and electro-physiological properties of synapses (Grunwald et al., 2001; Henderson et al., 2001; Takasu et al., 2002; Murai et al., 2003).
The diverse biological functions of EphRs and ephrins can be attributed to their binding properties as well as their ability to initiate bidirectional signaling between cells expressing receptors and cells expressing ligands (Davis et al., 1994; Holland et al., 1996; Himanen and Nikolov, 2003). Membrane localization of both proteins is essential for signaling via cell–cell contact. Upon binding, the EphR and ephrin, on respective cells undergo dimerization and clustering, which can be mimicked by artificial clustering of tagged soluble extracellular domains of either EphR or ephrin, leading to either receptor of ligand activation (Davis et al., 1994; Wilkinson, 2000; Martinez and Soriano, 2005). In mammals, studies have suggested that in adult CNS, ephrins are mostly expressed at the pre-synaptic terminals, while EphRs are clustered at postsynaptic membrane sites (Martone et al., 1997; Buchert et al., 1999; Contractor et al., 2002; Murai et al., 2003). EphA proteins are thought to change synaptic efficacy by restructuring synaptic connectivity through changes in dendritic spine morphology (Murai et al., 2003), while EphB proteins are thought to function through the regulation of the NMDA receptor neurotransmission (Murai and Pasquale, 2004).
EphR and ephrin proteins have been cloned from several invertebrate species including Drosophila melanogaster (Scully et al., 1999; Bossing and Brand, 2002) and the moth, Menduca sexta (Kaneko and Nighorn, 2003). They function in axonal pathfinding in the developing olfactory system of the moth (Kaneko and Nighorn, 2003) and in the establishment of topographic axonal projections in the developing visual system (Dearborn et al., 2002) and the mushroom body neurons (Boyle et al., 2006). Furthermore, in the Drosophila, the single EphR remains expressed in synaptic regions between retinal axons and medulla cells, beyond the developmental period, suggesting that in insects, also, EphR may have a role in synapse organization and function in the adult (Dearborn et al., 2002).
The aim of this study was to investigate the roles of the EphR and ephrin signaling in the developing and the mature brain of the honeybee, Apis mellifera (Am). We combined in situ hybridization and in situ labeling with Fc fusion proteins to determine expression patterns (for review see Wilkinson, 2000; Kaneko and Nighorn, 2003; Martinez and Soriano, 2005; Scalia and Feldheim, 2005; Boyle et al., 2006), at pupal and adult stages of development. The pattern of AmEphR and Amephrin expression at pupal stages suggested a role in brain development. In adult brain the expression of the AmEphR protein was confined to synaptic regions. This result coupled with evidence that Eph molecules affect behavior and support memory acquisition and retention in mammals (Gerlai, 2001) led us to examine whether EphR and ephrin signaling in the honeybee has a similar function. We used EphR-Fc and ephrin-Fc proteins to disrupt signaling (Gao et al., 1998; Gerlai et al., 1999; Dalva et al., 2000; Gerlai, 2001; Grunwald et al., 2001; Takasu et al., 2002; Murai et al., 2003). To evaluate the effects we used the proboscis extension reflex (PER) conditioning, which has been used extensively to study behavioral and physiological mechanisms of olfactory learning (Bitterman et al., 1983: Menzel, 1990, 2001; Maleszka et al., 2000; Farooqui et al., 2003). We show that inhibition of EphR/ephrin signaling leads to a reduction in the formation of long term memory and implicate EphRs and ephrin in the recall step in the memory pathway in honeybee.
METHODS
Honeybees
Worker honeybees were raised in hives located at the Australian National University, Canberra. For this study, larvae were used for RNA analysis and were selected randomly to represent prepupal stage of RNA expression. The head end of individual larvae (four samples per RNA preparation times three separate RNA preparations) was dissected for RNA preparations. Pupae (P1–3) were designated to one of the nine stages as per developmental criteria summarized in Ganeshina et al. (2000; see Table 1). To collect newly emerged (NE) adult bees, a single brood frame was obtained from the hive and placed in an incubator at 32°C (80% humidity). Individual insects were collected within 1–5 min after emerging from their cell and were either snap-frozen in liquid nitrogen or collected and used immediately for dissection of fresh tissue or caged and placed in an incubator maintained at 32°C (80% humidity) and used at days 4, 6, 8 (4d, 6d, 8d) postemergence. Forager bees were captured near the hive entrance and either snap-frozen in liquid nitrogen or caged briefly for transportation to the laboratory. To ensure that fully matured individuals were harvested, only those that carried pollen or nectar were collected. We estimate their age to vary from 21 to 35 days.
Sequencing and Annotation of AmEphR and Amephrin cDNAs
Two expressed sequence tags (ESTs) encoding EphR (accession numbers BI510551 and BI507081) were found by searching the collection of the honeybee brain ESTs available via the National Center for Biotechnology Information (NCBI) web server using the sequence for the Drosophila EphR protein. These ESTs were subsequently used to search the honeybee genome assembly V 2.0 available at the Baylor web site (www.hgsc.bcm.tmc.edu/projects/honeybee). The genomic scaffold designated Group14.2 that harbors the entire AmEphR gene was used to build a model structure for this gene. The annotation of the EphR gene was a two-step process involving ab initio predictions of intron/exon junctions followed by PCR amplification of DNA fragments corresponding to each predicted junction. The resulting cDNA sequence has been deposited in Gen-Bank (accession number AY921579).
As in the case of AmEphR, two ESTs encoding a putative ephrin were found in the honey bee brain library (accession numbers BI508521 and BI509243). However, since our annotation of the honey bee ephrin gene and its product (Amephrin) generated a protein similar to that predicted by NCBI, the NCBI entry (accession number XP_392239) will thereafter be referred to as Amephrin.
RNA Isolation, Electrophoresis, Blotting, and Hybridization
Detailed experimental protocols have been published previously (Kucharski and Maleszka, 2002, 2003). Preparation of brain tissue was carried out under permanent liquid nitrogen cooling as described previously (Kucharski and Maleszka, 2003). Total RNA was isolated using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA), followed by mRNA purification on Oligo (dt)25 magnetic beads (Dynal, Oslo). RNA samples were resolved using the glyoxal based system as per manufacturer’s instruction manual (Ambion). Following electrophoretic resolution the gels were quantified with the Vistra FluoroImager and blotted onto Hybond N+ nylon membranes (Amersham). Hybridization was carried out at 68°C (ExpressHyb solution, Clontech) for 16 h using P32-labeled probes (Redi-Prime kit, Amersham). Following posthybridization washes blots were exposed to a phosphorstorage screen (Molecular Dynamics, MD) and computer generated images (MD Phosphor-Imager 400S) of individual gels were analyzed using ImageQuant software.
Reverse Transcription-PCR for Analysis of Gene Expression
The presence of mRNA encoding AmEphR and Amephrin proteins in the whole head (larvae, P1, P2, P3) or brain (NE, 4d, 8d and foragers) was analyzed by reverse-transcription (RT)-PCR. Total cellular RNAs were extracted from fresh tissue using a single-step protocol for RNA isolation (‘‘RNAzol B,’’ Tel-Test, Friendswood, TX) as described previously (Vidovic et al., 1999; Osborne et al., 2002). Data were obtained from three separate RNA preparations at each age. The first-strand cDNA pool was prepared following a previously published protocol (Faber et al., 2005).
Oligodeoxynucleotide primers were designed from published sequences of the honeybee genome to amplify fragments of AmEphR, predicted PCR fragment of 536 base pair (bp) and Amephrin, predicted size of 500 bp. PCR amplifications were performed in a 20 μL reaction volume using standard techniques (Faber et al., 2005).
In Situ Hybridization
Whole heads from P1–3 and brains from anesthetized NE and forager bees were dissected into phosphate-buffered saline (PBS; pH 7.4) and immersed in 4% paraformaldehyde (PFA) in PBS for 4 and 2 h, respectively, at room temperature. Tissue was then transferred to 18% sucrose in PBS and sections were cut on a cryostat at a thickness of 20 μm. Digoxigenin (DIG)-labeled riboprobes were synthesized using a DIG RNA Labeling Kit (Roche, Indianapolis, IN), following the manufacturer’s protocol, from a plasmid construct encoding a 460 and 469 bp sequence of the AmEphR and Amephrin gene, respectively. Localization of the mRNA transcripts was achieved by incubating air-dried tissue sections in PBS for 5 min, postfixing in 4% PFA for 20 min and rinsing once in PBS with 0.1% active diethylpyrocarbonate (DEPC; ICN biomed) for 10 min to inactivate RNases. Tissue was then washed in PBS and incubated in 5 μg/mL Proteinase K (Sigma) in PBS at 37°C for 10 min then rinsed once in PBS with 0.1% DEPC for 5 min and dehydrated with increasing concentrations of ethanol and air dried for 20–30 min. Hybridization was carried out at 55°C overnight in a prehybridization buffer described in Marotte et al. (2004). We have examined sections that represented all the major brain regions (4 sections per probe from each brain, from 4 individual brains at P1–3, NE, 8d and forager worker honeybee). Furthermore, at each time point examined, we included adjacent sections from the same brain to check probe specificity by hybridizing with the sense probe [Fig. 2(C,D,G,H)]. Post-hybridization protocol, including the detection of bound DIG probe, was done as described by Marotte et al. (2004). Individual images were viewed with a Zeiss Axioskop microscope and digitally photographed using Spot advanced software and edited as required for brightness and contrast using Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA).
Figure 2.

Analysis of AmEphR and Amephrin transcripts in the honeybee brain using in situ hybridization. Localization of AmEphR (A,E) and Amephrin (B,F) mRNAs in pupae (A–D) and adult brain (E–H). Adjacent sections were incubated with sense riboprobes for AmEphR (C,G) and Amephrin (D,H). (A,B) An overall view of whole-brain frontal section showing EphR and ephrin expression, respectively, in the cell body rind in the mushroom body (MB) and the optic lobes (OL). (E,F) Similar expression patterns were found in the adult. In the MB, expression was detected in the Kenyon cell body regions inside (arrows) and outside the calyces (arrow heads). In the optic lobe, mRNAs for the EphR and ephrin was detected in rows of cells located between the lamina (la) and the medulla (me) and between the medulla (me) and the lobula (lo). Scale bars, 0.5 mm.
In Situ Labeling with Fc Fusion Proteins
Fc fusion proteins of Moth, Manduca sexta (Ms), EphR (EphR-Fc) and ephrin (ephrin-Fc) (the extracellular domain of Ms-EphR or Ms-ephrin linked to human IgG-Fc, Kaneko and Nighorn, 2003; Boyle et al., 2006), and IgG-Fc (Jackson ImmunoResearch, West Grove, PA) were reacted with unfixed cryosections (20 μm) of honeybee brains from bees aged P1, NE bees, and foragers. In situ labeling was carried out as described in Kaneko and Nighorn (2003), with following modifications. Incubations with Ms-EphR-Fc, -ephrin-Fc, or IgG-Fc (12 μg/mL) were for 2–4 h at room temperature, followed by 5 washes in PBS for 7 min each. Reacted samples were then counter stained with 4′,6-diamidino-2-phenylindole (DAPI). Additional sections were stained with cresyl fast violet to show morphological features. Individual images were viewed with a Nikon Opti-phot Epifluorescence microscope and digitally photographed using Spot advanced software. Editing of digital images was done using Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA) for the purpose of contrast and color enhancement and to combine images from tissue that were labeled with two fluorescent markers.
Injections of Fc Fusion Proteins into the Brain and Conditioning and Testing Procedures
Young adult bees (6d) were anesthetized on ice and mounted in thin-walled aluminum tubes (7 mm in diameter) such that only the antennae and mouthparts were free to move. Individuals were fed with 1M sucrose solution and kept overnight in a dark humid space. On the following day, individual bees were injected with either MsEphR-Fc or Msephrin-Fc (unclustered forms at 0.27 μg/μL, diluted in bee ringer). Controls received the same volume of IgG-Fc (0.27 μg/μL diluted in bee ringer) or bee ringer. Prior to each injection, the lens covering the median ocellus was removed creating a hole and reagents (0.25 μL) were delivered into the brain using a 25-μL Hamilton syringe with a repeating dispenser. This approach of reagent delivery allowed for rapid access to the whole brain (Dacher et al., 2005). Training sessions began either one hour before or one hour after the injections. The learning protocol used in this study was an adaptation of the olfactory conditioning of the PER (Menzel, 1990; Maleszka et al., 2000; Maleszka and Helliwell, 2001).
For olfactory conditioning, each bee was allowed to first smell the rewarding stimulus (limonene, 4 μL/mL in 1M sucrose) for 5 s, followed by the application of the reward stimulus (a droplet of limonene, 4 μL/mL in 1M sucrose) on the antenna. This resulted in the PER and the tasting of the sugar reward. Each bee was then presented with the nonrewarding stimulus (natural vanilla, 4 μL/mL, in saturated NaCl). Each trial was either done once (short term memory) or repeated three times at 6-min intervals (long term memory). The test for the retention of associative learning was carried out 1 h (short-term) and 24 h (long-term) after the training session was completed.
First, the nonrewarding and then the rewarding stimulus was presented to each individual, and the presence or absence of the PER was noted. Bees that withheld their proboscis when presented with the nonrewarding stimulus (vanilla), and then extended the proboscis when presented with the rewarding stimulus (limonene) were scored as having responded correctly. Bees responding to the nonrewarding stimulus or to both stimuli were recorded as responding incorrectly. For statistical analysis, the data were compared using the Pearson’s χ2 test with Yates’ continuity correction. p-values were computed by Mote Carlo simulation. All calculations were done with the GNU/R software.
RESULTS
Characterization of the AmEphR and Amephrin Genes and Their Products
In the honeybee genome assembly version 2.0, we have identified the genomic loci that encode putative EphR (AmEphR) and ephrin (Amephrin) proteins. The AmEphR genomic locus is at least 33 kb, but contains largely intronic sequences that span 90% of the gene. Eleven introns ranging in size from 73 to 19,171 bp have been confirmed in the coding region. The open reading frame (ORF) encodes the AmEphR protein of 996 amino acids while the Amephrin ORF encodes a predicted protein of 342 amino acids. Interestingly, the sizes of each ORF are substantially smaller than the mRNA transcripts identified in Northern analyses indicating that only part of each messenger RNA seen in the northern blot are translated [Fig. 1(E,F)]. However, as both the AmEphR and Amephrin ORFs are flanked by multiple in-frame and out-of-frame stop codons, the cDNAs almost certainly include the entire ORFs (GeneBank accession nos: AY921579 and XP_392239.1).
Figure 1.
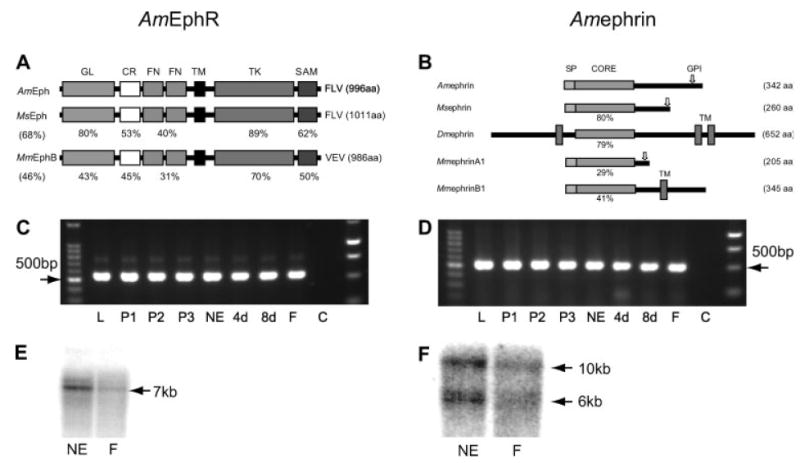
Schematic diagram of EphR and ephrin proteins and mRNA expression of AmEphR and Amephrin in developing and adult brains. (A) Diagram comparing the domain structure of AmEphR with MsEphR and mouse, MmEphB2R. Domain structure from N terminus: GL, globular domain; CR, cystein-rich domain; FN, fibronectin type-III repeats; TM, transmembrane motif; TK, protein tyrosine kinase domain; SAM, sterile α motif. Amino acid similarities between equivalent regions are presented as percentages. The overall similarity is indicated in parentheses. (B) Schematic diagram comparing Amephrin with Msephrin, Dmephrin (Dephrin from Drosophila melanogaster) and mouse (Mus musculus), MmephrinA1 and MmephrinB1. The relevant segments are indicated as secretion signal peptides (SP), conserved ephrin core domains (CORE), transmembrane domains (TM), and GPI-anchoring domain (arrow). Amino acid similarities between equivalent ephrin core domains are presented as percentages and the size of each protein is indicated by the number of amino acids (aa). (C,D) AmEphR and Amephrin mRNA expression in the developing and adult brains. Agarose gel electrophoresis of 8 μL of PCR product. A band of 536 bp represents AmEphR (C) and 500 bp Amephrin (D). PCR products for each gene, defined by specific set of primers, were amplified from the same cDNA pool at each age. Different ages are designated by L, larvae; P1–P3, pupae day 1, 2, 3; NE, newly emerged; 4d, 8d, 4 and 8 days postemergence; F, foragers; C, control that did not contain RNA. DNA size markers, 100 bp and 1 kb (Promega, Madison, USA). (E) Northern blot analysis shows that mRNA extracted from honeybee brain at two ages and hybridized with random-primed specific probes for AmEphR and Amephrin recognize a single transcript (kb) for the EphR (E) and two for the ephrin (F) at two ages, NE, newly emerged and F, foragers.
The N-terminal sequences of AmEphR contain the globular domain or the ligand binding domain, a cysteine-rich region, and two fibronectin type III segments near the transmembrane motif. These show 80, 53, and 40% amino acid identity, respectively, with MsEphR (Kaneko and Nighorn, 2003) motifs [Fig. 1(A)]. The intracellular domain is characterized by a juxtamembrane region that sits between the tyrosine kinase catalytic domain and the transmembrane motif. The kinase amino acid sequences show 89 and 70% identity with the MsEphR (Kaneko and Nighorn, 2003) and the mouse EphB2 receptor, respectively [Fig. 1(A)]. The carboxy terminal tail of the AmEphR contains the sterile α motif (SAM). The last three C-terminus residues, FLV, fit the consensus motif of F-x-V/I that binds to one group of PDZ domain-containing proteins such as p55, Tiam-1, and AF-6 (Songyang et al., 1997). Like the Manduca and Drosophila proteins, the AmEphR is equally similar to the mammalian Eph family A and B subclasses, both overall and in each domain, further supporting the idea that the insect receptors may represent a prototypical member of the Eph family (Scully et al., 1999).
The sequence analyses of Amephrin predict an N-terminal secretion signal peptide and a GPI-anchoring motif near the C terminus [Fig. 1(B)]. The central region of Amephrin contains the conserved ephrin core domain, believed to mediate receptor binding. This shares highest homology with the central regions of Manduca sexta (80%) and Drosophila (79%), and less with the mammalian ephrins-A1 (29%) and eph-rin-B1 [41%, Fig. 1(B)]. Likewise, the overall structure of Amephrin protein is more similar to Msephrin than to known ephrins in 3 dipteran species, D. melanogaster, D. pseudoobscura, and Anopheles gambiae. Amephrin has a putative GPI-anchoring motif, whereas the dipteran ephrins are predicted to be transmembrane proteins (Maleszka, unpublished observations; Bossing and Brand, 2002). Also, Amephrin appears not to have the N-terminal extension of ~200 amino acids present in both Drosophilae. Interestingly, this extension is not a feature of dipteran ephrins as it is not found in the mosquito (Maleszka, unpublished observations). While the core domain of Amephrin is more similar to mammalian B-subclass ephrins, its overall structure as a GPI-anchored protein resembles the ephrin-As [Fig. 1(B)].
AmEphR and Amephrin mRNA Is Strongly Expressed in the Brain During Pupal Development
To demonstrate the temporal and spatial expression of the two genes, we analyzed mRNA from developing and adult honeybee tissue by RT-PCR [Fig. 1(C,D)], Northern blots [Fig. 1(E,F)], and in situ hybridization (Fig. 2). RT-PCR amplification products of the expected sizes for the AmEphR [536 bp; Fig. 1(C)] and Amephrin [500 bp, Fig. 1(D)] genes were readily detectable in the total RNA extracted at various stages of development from the tissues of the whole head [L, P1–3; Fig. 1(C,D)] and brain only [NE, 4d, 8d, F; Fig. 1(C,D)]. Both genes were expressed in the honeybee head during the larval and early pupal development and in brains of NE bees and older foragers. Northern blot analyses using brain mRNA extracted from NE and forager bees show the expression of one EphR mRNA transcript (~7 kb) in the NE adult and foraging honeybee [Fig. 1(E)] brains while two mRNA transcripts (6 and 10 kb) represented the expression of the Amephrin gene [Fig. 1(F)]. As homology searches of the honeybee genomic sequence data base failed to reveal any additional Amephrin-related genes the two mRNA transcripts for Amephrin suggest the existence of at least 2 splice variants.
In situ hybridization of specific riboprobes to brain tissue sections of pupal and adult individuals was done to determine the cellular expression of mRNA for AmEphR and Amephrin (Fig. 2). Honeybee brain structure is characterized by a clear separation of the cell soma rind (comprising neurons and glia) and the neuropile. The underlying neuropile is devoid of neuronal somata but contains neuronal processes and glial cell bodies (Hähnlein and Bicker, 1996). Widespread expression of both mRNA transcripts was readily detected in the cell body regions of the developing and adult honeybee brain (Fig. 2). At P1, a period of continued prolific neurogenesis in the honeybee brain, an intense hybridization of AmEphR [Fig. 2(A)] and Amephrin [Fig. 2(B)] riboprobes was evident in the somata associated with the developing optic lobes and mushroom bodies. Neurones associated with the developing antennal lobe (not shown) and cells posterior to the antennal lobe expressed mRNA for both genes. In all regions mRNA expression for both receptor and ligand appeared to be present in all cells (excluding glial cells in the neuropile), resulting in diffuse but intense staining, with no differential hybridization demonstrated between the two genes in the different neuronal populations within the soma rind. Similar expression patterns for both genes were seen at P2–3 (data not shown). No significant hybridization was seen in sections incubated with the sense probe [Fig. 2(C,D)]. The uniform staining for both genes across the cell body regions of the brain during early pupal development suggests that most, if not all, neurons, at different stages of neurogenesis and differentiation, express mRNAs for both genes.
In the adult honeybee brain, which begins at the NE stage and marks the end of neurogenesis in most regions, expression of mRNA for EphR [Fig. 2(E)] and ephrin [Fig. 2(F)] was most evident in cell body regions of the mushroom bodies and the optic lobes. In the mushroom bodies the Kenyon cell somata that reside in [Fig. 2(E), arrows] and around [Fig. 2(E), arrow heads] the calyces were equally labeled for EphR expression. A similar pattern (albeit at a lesser intensity) of mRNA expression for Amephrin to that of AmEphR is illustrated in Figure 2(F). Similarly, in the brains of foragers (data not shown) the expression of Amephrin gene was at a reduced level compared to the receptor gene. No significant hybridization was seen in sections incubated with the sense probe [Fig. 2(G,H)].
Identification of Sites of Ligand and Receptor Protein Binding
To understand the possible functions of EphR and ephrin signaling in the developing and adult honeybee brain, it is important to determine where these interactions occur at the protein level. We used recombinant proteins as probes, consisting of the extracellular domain of the MsEphR to localize Amephrin expression and Msephrin for AmEphR localization. The high degree of overall sequence similarity between the MsEphR and AmEphR (68%) and Msephrin and Amephrin (80% in the central region believed to mediate receptor binding) allowed the use of these proteins. Analysis of the distribution of the two proteins in the developing (P1–3) and adult (NE) brain uncovered differences in their expression patterns.
AmEphR and Amephrin Protein Expression Patterns in the Developing Mushroom Bodies
The morphological development of the honeybee mushroom bodies has been fully described previously (Malun, 1998; Farris et al., 1999; Kurshan et al., 2003). Briefly, at P1–2 mushroom bodies are clearly defined paired structures [Fig. 3(A,B)] in the proto-cerebrum of each brain hemisphere [Fig. 3(A)]. They consist of undifferentiated clusters of neuroblasts [N, Fig. 3(B)] and differentiated Kenyon cells (Farris et al., 1999). Kenyon cells arise from the larger cells located within the clusters [Fig. 3(B), see also Figs. 1 and 2 in Ganeshina et al. 2000] and occupy regions around the neuroblast clusters [Fig. 3(A,B)]. In the developing brain of the honeybee, compact clusters of neuroblasts that give rise to the mushroom bodies have been identified as early as the first larval stage of development (Malun, 1998; Farris et al., 1999) and the production of cells, primarily the Kenyon cells, continues to mid pupal stage (Farris et al., 2004). Kenyon cell axons give rise to the peduncular neuropile [P, Fig. 3(B,D,G,J)] while the calycal neuropile [Ca, Fig. 5(A)] contains the dendritic projections of the Kenyon cells and the innervating pre-synaptic terminals. Both neuropiles begin to develop before the prepupal stage and occupy regions in the mushroom body that are entirely distinct from the Kenyon cell somata (Farris et al., 1999).
Figure 3.

Distribution of the AmEphR and Amephrin proteins in the mushroom body during early pupal development. (A) Cresyl fast violet-stained frontal section of the honeybee brain at P1, showing mushroom bodies (MB) in relation to other structures. Optic lobes (OL), medulla (me) and lobula (lo). Scale bar, 0.5 mm. (B) Magnified view of the boxed area in A, showing the medial (left, boxed) and lateral developing calyces of the MB. Each neuroblast (N) cluster is surrounded by darkly stained Kenyon cell bodies. (C) AmEphR expression (arrows) was detected by in situ labeling with ephrin-Fc on cryosections of fresh tissue. (F) Amephrin expression (arrow heads) was detected by in situ labeling with EphR-Fc. (D,G,J) Chromosomal DNA was counterstained with DAPI (depicted as purple in the composite images). Peduncular neuropile (P). (E,H) Composite images place the EphR expressing cells (depicted as yellow/orange) in a region occupied by the most recently born Kenyon cells and the Amephrin expressing cells within the neuroblast cluster, respectively. (I–K) No immunofluoresence was detected in sections stained with IgG protein. Scale bar in K = 100 μm in C–K. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 5.
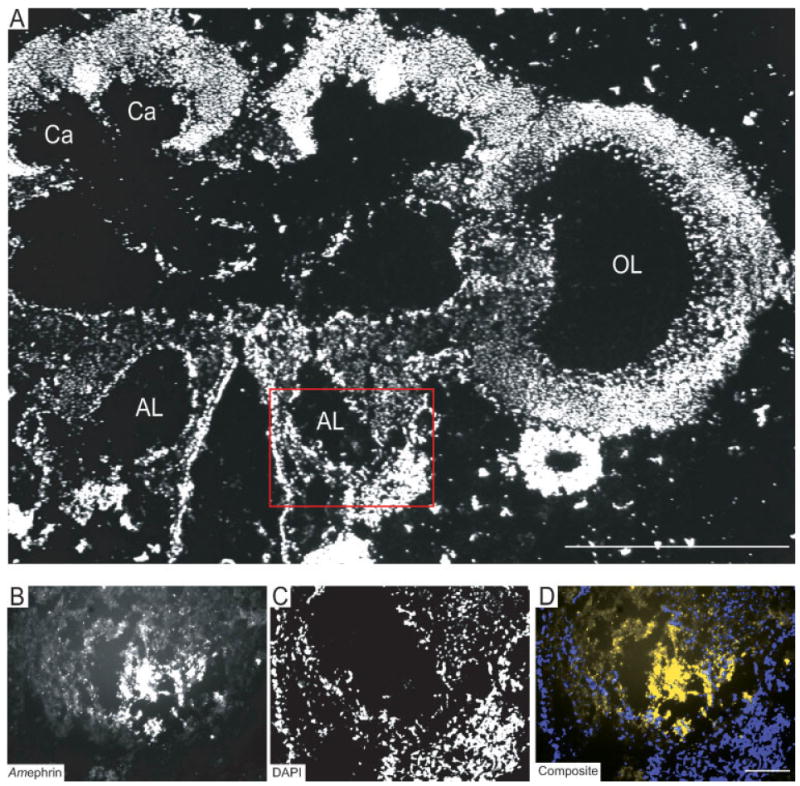
Localization of the Amephrin protein in the antennal lobes of the honeybee brain during early pupal development. (A) DAPI-stained frontal section of the honeybee brain at P1, showing the optic lobe (OL) Calyx (Ca) of the mushroom bodies, Antennal lobes (AL). Scale bar, in A = 100 μm. Red box denotes area of higher magnification and ephrin protein expression in B–D. (B) Amephrin protein expression was detected by in situ labeling with EphR-Fc. (C) Chromosomal DNA was counterstained with DAPI. (D) Composite image (depicted as yellow) demonstrated that Amephrin protein was expressed in the neuropile of the AL. Scale bar, in D = 100 μm in B–D. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.] subcompartments, collectively known as the olfactory glomeruli, surrounding a central core of course-textured neuropile [Fig. 6(G–I); see also Fig 1, Brown et al., 2002]. Strong staining for AmEphR was detected in glomeruli positioned ventrally in the antennal lobe [Fig. 7(G), arrows] with less intense staining in the cortex of glomeruli dorsally [Fig. 7(G), arrow heads].
During early pupal development (P1–2), in sections reacted with the ephrin-Fc affinity probes, cells with weak immuno-reactivity were visible as narrow strips of punctate fluorescence on both sides of the neuroblast clusters of the mushroom bodies [Fig. 3(C), arrows]. Counter-staining with DAPI, for localization of DNA, confirmed that the immunolabeling was con-fined to the cellular region [Fig. 3(D)]. The resultant composite image [Fig. 3(E)] suggests that the expression of EphR, as demonstrated by the binding of ephrin-Fc protein, was most likely associated with cells located in a region shown by Farris et al. (1999) to contain the most recently born Kenyon cells. It is worth mentioning that in the absence of a specific label to identify glial cells, some of the AmEphR expressing cells identified around the neuroblast clusters could include newly born glial cells that only transiently express AmEphR protein.
Interestingly, in the sections reacted with the EphR-Fc affinity probes the most conspicuous immunofluorescence was seen as a cluster of intensely labeled cells in the regions of the neuroblast cluster in the mushroom body [Fig. 3(F), arrow heads]. The composite image places the Amephrin protein expressing cells within the neuroblast cluster [Fig. 3(H)]. No staining of the mushroom body neuropile [P, Fig. 3(C,D,F,G)] for either of the two proteins was observed at the pupal stage examined. Importantly, no immunofluoresence was detected in sections stained with IgG protein [Fig. 3(I–K)].
AmEphR and Amephrin Protein Expression Patterns in the Developing Optic Lobes
Analysis of the developing optic lobes at P1–2 [Fig. 4(A)] revealed that the most striking immunolabeling for both AmEphR and Amephrin proteins was in the neuropile regions, rather than in regions occupied by cell bodies [Fig. 4(B–G)]. The optic lobes, which process visual information received from the photoreceptors of the compound eye, comprise three separate neuropile regions: the lamina, medulla, and lobula [Fig. 4(A), see also Mobbs, 1982; Ehmer and Gronenberg, 2002]. Figure 4(B) shows high EphR expression in the lobular neuropile. Counter-staining with DAPI [Fig. 4(C)] confirms that expression is absent in the surrounding cell soma region [Fig. 4(D)]. Amephrin protein expressing neurites were also visible in regions of the lobula [Fig. 4(E–G)] and were in a complementary pattern to those expressing EphR protein [Fig. 4(B–D)]. Regions of the neuropile of the lobula with high EphR protein expression showed little or no ephrin staining, and vice versa.
Figure 4.
Localization of the AmEphR and Amephrin proteins in the optic lobes of the honeybee brain during early pupal development. (A) Cresyl fast violet-stained frontal section of the honeybee brain at P1, showing the optic lobes (medulla, me; lobula, lo) in relation to other structures such as Mushroom bodies (MB). Scale bar, 0.5 mm. (B) AmEphR expression (arrows heads) was detected by in situ labeling with ephrin-Fc on cryosections of fresh tissue. E, Amephrin expression (arrow heads) was detected by in situ labeling with EphR-Fc. (C,F) Chromosomal DNA was counter-stained with DAPI. (D,G) Composite images demonstrate that AmEphR and Amephrin were expressed in a complementary pattern in the neuropile of the lobula (lo). Scale bar in G = 100 μm in B–G. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
AmEphR and Amephrin Protein Expression Patterns in the Developing Antennal Lobes
Use of EphR and ephrin-Fc affinity probes only detected the presence of the Amephrin protein in the region of the developing antennal lobes [Fig. 5(A–D)]. It was present largely, but not exclusively, in neuropile region [Fig. 5(B)] as confirmed by counter-staining with DAPI [Fig. 5(C)]. The resultant composite image [Fig. 5(D)] suggests that the expression of Amephrin protein, as demonstrated by the binding of EphR-Fc protein, was associated with neurites located in the region of the developing neuropil of the antennal lobe. AmEphR protein expression (data not shown) was not detected in the antennal lobe.
AmEphR Protein Expression Patterns in the Mature Brain of the Honeybee
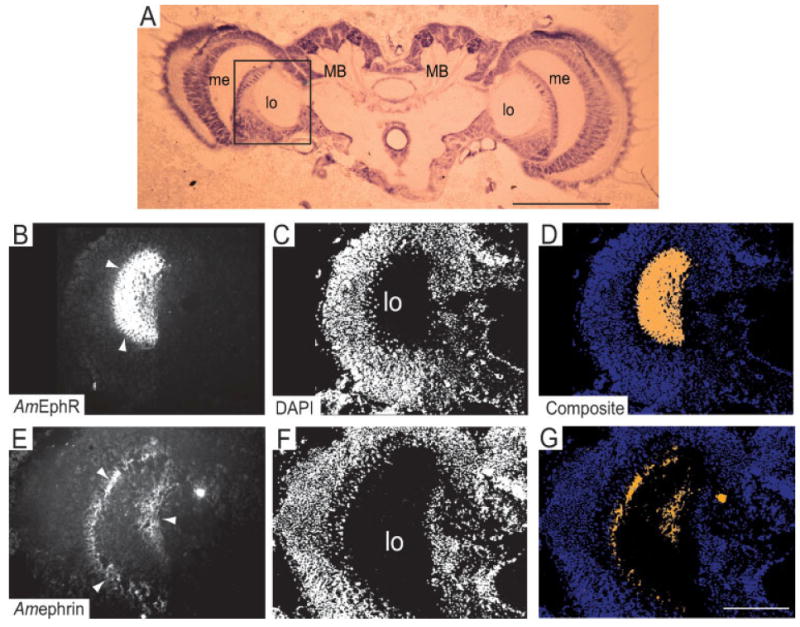
In NE young adult honeybees, use of EphR and ephrin-Fc affinity probes only detected the presence of the EphR protein in the brain. It was present in the neuropiles of the mushroom bodies and the central body [red boxed region, Fig. 6(A)] and the optic [white boxed region, Fig. 6(A)] and antennal lobes [blue boxed region, Fig. 6(A)]. In the mushroom body, strong staining for EphR was detected in the medial lobe (Strausfeld et al., 2000) also known as the β lobe [Fig. 6(B–D)]. These lobes are sites of synaptic contacts in which Kenyon cell axons project and form pre- and postsynaptic contacts with processes of extrinsic neurons (Strausfeld et al., 2000; Farris et al., 2004). AmEphR was also detected in the two small circular structures, the noduli [Fig. 6(E–G), positioned at the rear of the central body region [CB, Fig. 6(A); see also Mobbs, 1982].
Figure 6.

Localization of the AmEphR protein in the mushroom body and the central body of the adult honeybee brain. (A) Cresyl fast violet-stained frontal section of the adult honeybee brain. Red box denotes area of higher magnification and EphR expression in B–G. White box denotes area of higher magnification and EphR expression in A–F (Fig. 7). Blue box denotes area of higher magnification and EphR expression in G–I (Fig. 7). Medulla, me; Lobula, lo, Antennal lobes (AL), Scale bar, 0.5 mm. (B,E) AmEphR expression was detected by in situ labeling with ephrin-Fc on cryosections of fresh tissue. (C,F) Chromosomal DNA was counterstained with DAPI (depicted as purple in the composite images). (D,G) Composite images demonstrate that AmEphR was expressed (depicted as yellow) by the neuropile in the lobes of the MB (arrows heads in B), specifically in the medial lobe, also known as the β lobe and in the noduli (n) respectively. Scale bar in D = 100 μm, in B–D and in G = 100 μm in E–G. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
In the optic lobes, both the medulla and the lobula [white boxed, Fig. 6(A)] show intense staining for the AmEphR [Fig. 7(A)]. Counter-staining with DAPI confirmed immunolabeling was confined to the neuropile region [Fig. 7(B)] and this is illustrated in the composite image [Fig. 7(C)]. In the medulla, the strong expression of EphR was most likely associated with neurites within the inner and outer regions [arrow heads in me, Fig. 7(A)]. These regions are separated by the middle layer, the serpentine layer, which contains axons and tangential neurons (Ehmer and Gronenberg, 2002). It is likely that some of the neurites labeled for the AmEphR within the dorsal aspect of the medulla belong to neuronal cells with axonal projections that connect the medulla and the mushroom bodies via the anterior superior optic tract. Their cell bodies are situated in a cluster between the dorsomedial edge of the medulla and the base of the lateral calyx [see Figs. 2(d) and 3(c) in Ehmer and Gronenberg, 2002]. However, the bulk of AmEphR protein detected in the optic lobes is likely to be expressed on fiber elements of visual neurons linking the lamina with the medulla and the medulla with the lobula (see Fig. 3 in Ribi and Scheel, 1981).
Figure 7.

Localization of the AmEphR protein in the neuropiles of the optic and antennal lobes. (A,G) AmEphR expression was detected by in situ labeling with ephrin-Fc on cryosections of fresh tissue. (B,E,H) Chromosomal DNA was counterstained with DAPI. (C,I) Composite images demonstrate that AmEphR was expressed in the medulla (me) and the lobula (lo, arrow heads in A) and the glomeruli, positioned in the ventral end of the antennal lobe (arrows in G) and in the cortex of glomeruli at the dorsal end of the anntenal lobe (arrow heads, G). (D–F) The Fc control probe produced no significant staining in the optic lobes or any of the other brain regions examined (data not shown). Scale bar in I = 100 μm (A–I). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The antennal lobe neuropile in adult honeybees [blue box, Fig. 6(A)] is structured into many spherical The cortex of the mature glomeruli is the site of synaptic contacts between the antennal sensory afferents, local interneurons, and output relay neurons (Hahnlein and Bicker, 1996). The output relay neurons project to the lip region of the calyx and to the lateral protocerebral neuropile (Gascuel and Masson, 1991). The composite image places some DAPI stained cell nuclei [purple in Fig. 7(I)] within the neuropile region and in regions surrounding the glomeruli, suggesting that some glial cells may be expressing EphR protein.
Surprisingly, no protein expression of Amephrin was detected in the brains of adult bees (data not shown). This is in contrast to the mRNA data from RT-PCR, Northerns and in situ hybridization that clearly demonstrate the presence of mRNA for ephrins in adult brains.
Blocking the EphR and Ephrin Signaling with EphR-Fc and Ephrin-Fc Results in Deficits in Associative Learning and Memory
EphR/ephrin-Fc fusion proteins have been valuable tools with which to study the role of Eph molecules on synaptic function, in vitro (Gao et al., 1998; Dalva et al., 2000; Gerlai, 2001; Grunwald et al., 2001; Takasu et al., 2002; Murai et al., 2003) and on learning behavior in mice (Gerlai et al., 1999; Gerlai, 2001). To investigate the effects of disruption of signaling by EphR and ephrin proteins on learning and memory, we used EphR-Fc or ephrin-Fc (unclustered form) for injecting into the brain of the honeybee. While tagged unclustered extracellular domains of receptor or ephrin can bind to their respective proteins on the cell surface, they do not trigger receptor/ephrin activation unless they are artificially clustered. We therefore expect the unclustered forms of EphR-Fc or ephrin-Fc proteins, injected into the brain of the honeybee to act as blockers by occupying endogenous receptor/ligand binding sites and preventing signal initiation. Young adults were injected once with either EphR-Fc or ephrin-Fc and controls, human IgG-Fc or Bee Ringer, 1 h before a three trial conditioning involving paired presentation of two odors. This could potentially affect both the acquisition and the subsequent consolidation of memory. The test for the retention of associative learning was analyzed 24 h after training by presenting first the nonrewarding and then the rewarding stimulus to each individual. Long-term memory is dependent on new protein synthesis and requires a set of receptors and transporters in specific parts of the brain.
Results in Figure 8(A) show the percentage of bees, from each group that responded correctly when presented with the dual stimulus. Long-term memory of PER conditioning was reduced in groups that were injected with EphR-Fc (29.1%) or ephrin-Fc (30.3%) proteins compared with IgG (50%) or Bee Ringer (56.6%) controls. Pairwise comparisons revealed a significant difference between the EphR-Fc treated (n = 55) and Bee Ringer treated (n = 53) groups (**p < 0.05; χ2 = 8.35; χ2 test) or EphR-Fc treated (n = 55) and hIgG-Fc treated (n = 70) groups (*p < 0.01; χ2 = 5.57; χ2 test). Similarly, there was a significant difference between the animals treated with ephrin-Fc (n = 66) and those treated with Bee Ringer (n = 53) groups (**p < 0.05; χ2 = 8.34; χ2 test) and between ephrin-Fc treated (n = 66) and hIgG treated (n = 70) groups (*p < 0.01; χ2 = 5.47; χ2 test).
Figure 8.

Effects of EphR-Fc, ephrin-Fc and Fc control on memory of an associative olfactory task. (A) Effects of EphR-Fc, ephrin-Fc and Fc control on long term memory, tested 24 h after training. The percentage of correct responses was evaluated by the PER conditioning. Long-term memory of PER conditioning was reduced in groups that were injected with EphR-Fc (29.1%) or ephrin-Fc (30.3%) proteins compared with IgG (50%) or Bee Ringer (56.6%) controls. (B) Tests for the retention of associative learning at a shorter time point relative to training. Individuals were injected with similar reagents 1 h before conditioning, with a single trial involving paired presentation of two odors. The retention of associative learning was carried out 1 h after training. Over 60% of subjects, in each of the two controls responded correctly to the conditioned odor compared to the 57.7% and 46.5% of subjects treated with EphR-Fc and ephrin-Fc. (C) The effect of preconditioning on short term memory recall. Individuals were conditioned, with a single trial dual scent, 1 h before treatment with ephrin-Fc (n = 66) and the hIgG-Fc (n = 76) control and tested 1 h after treatment. Over 77% of control group responded correctly compared with 31.8% of correct responses in the ephrin-Fc treated group.
To test for the retention of associative learning closer to the time of training or short-term memory, lasting for a few hours, we treated individuals with similar reagents 1 h before conditioning with a single trial involving the paired presentation of two odors. The test for retention of associative learning was carried out 1 h after the training session was completed by presenting first the nonrewarding and then the rewarding stimulus to each individual, and noting the presence or absence of PER. Results in Figure 8(B) show that over 60% of subjects, in each of the two controls, responded correctly to the conditioned odor compared to 57.7% and 46.5% of subjects treated with EphR-Fc (n = 52) and ephrin-Fc (n = 71), respectively. Memory retention was also significantly reduced in the ephrin-Fc treated group when compared with the hIgG-Fc treated (n = 74) group (*p < 0.01; χ2 = 5.74; χ2 test) and the Bee Ringer treated (n = 42) controls (**p < 0.05; χ2 = 3.35; χ2 test). However, the slight reduction in short term memory retention after treatment with EphR-Fc was not significantly different from the two controls.
To examine the effect of ephrin-Fc on short term memory recall, individuals were conditioned, with a single trial dual scent, 1 h before treatment with ephrin-Fc (n = 66) and the hIgG-Fc (n = 76) control. One hour after treatment, subjects were tested for the retention of associative learning. Figure 8(C) shows the percentage of correct responses for the two groups. In the hIgG-Fc treated control group, over 77% of individuals responded correctly compared with 31.8% of correct responses in the ephrin-Fc treated group. A pairwise comparison revealed a significant difference between the ephrin-Fc and hIgG-Fc treated groups (**p < 0.05; χ2 = 30.1; χ2 test), indicating that experimental amnesia was also induced by post-training injections with ephrin-Fc.
The point of using both paradigms is to determine whether the experimental treatment affects the long term memory cascade or transient modifications underlying short term memory. For each of the three sets of data (Fig. 8(A–C)], we performed at least three replications in which the investigator was blind to the treatment condition until data from each trial was collected and analyzed. Furthermore, no toxic side effects in a form of either mechanical or neurotoxic lesions, which could account for the amnesic effect, were produced by the injection of Fc- fusion proteins at the described concentrations. Histological examination of the brain tissue (results not shown) of injected animals showed no observed lesions as a consequence of injections at least no different to the controls, and therefore does not affect either learning or retention. The survival rate of EphR/ephrin-Fc groups of treated bees was indistinguishable from that of control-injected bees.
DISCUSSION
We have identified the honeybee homologs of EphR (AmEphR) and its ligand (Amephrin) and report on their expression and distribution at the mRNA and protein levels in the brain of pupal and adult bees. Our observations show that most neurons, at different stages of neurogenesis as well as postmitotic neurons, express mRNAs for both genes. However, protein expression is regulated. Their restricted expression in the mushroom bodies, during pupal development, supports a role in progenitor cell function. In the adult brain, the presence of AmEphR protein in regions of the neuropile supports a potential function in synaptic plasticity. This is supported by effects of in vivo disruption of EphR/ephrin signaling on olfactory learning and memory and implicates EphRs and ephrin in the recall step in the memory pathway.
The significance of the different patterns of AmEphR and Amephrin expression at the mRNA and protein levels is not clear. However, we suggest that the differential expression of the two gene products occurs at the level of protein synthesis. This is supported by our observation in the developing mushroom bodies where neuroblasts retain the expression of Amephrin protein while newly produced neurons, the Kenyon cells, up regulate the expression of AmEphR protein as they appear around the proliferative zone. The oldest cells are pushed away from the center of the calyx (Farris et al., 1999). The most recently generated neurons remain in the position occupied by the outgoing neuroblasts (Farris et al., 2004), when the neuroblasts undergo programmed cell death during the mid-pupal stage (Ganeshina et al., 2000).
The complementary expression of Amephrin protein in the neuroblast and AmEphR protein in the newly formed cells points to a role for these molecules in progenitor cell function and or differentiation of Kenyon cells. Indeed, expression of EphRs/ephrins, by cortical precursors during early stages of rodent brain development suggests they function in neurogenesis (Aoki et al., 2004; Holmberg et al., 2005; Depaepe et al., 2005). Ephrins and their receptors can mediate repulsive interactions and may function in the developing mushroom bodies to restrict intermingling between neuroblast and the newly differentiated cells. Furthermore, EphR/ephrin activation may be instrumental in the induction of initial process elaboration by Kenyon cells following soon after the neuroblasts exit the cell cycle as was suggested for early cortical precursors in rodent brain development (Gao et al., 2000; Zhou et al., 2001).
While our observations suggest that AmEph/ephrin systems may not regulate the development of the expanding neuropile in the developing mushroom bodies, the prevalence and intensity of AmEphR and Amephrin protein expression in the optical lobe and Amephrin protein in antennal lobes indicates their presence on the developing neuronal processes of neurons associated with vision and olfaction. Their expression coincides spatially and temporally with rapid growth and compartmentalization of the honeybee brain neuropile (Kurshan et al., 2003). The spatial distribution of AmEphR and ligand in the optical lobes, being restricted to separate regions of the neuropile in a complementary pattern, favor a repellent rather than an attractive interaction between these molecules. The repulsive effects resulting from EphR-ephrin activation have been shown to regulate the guidance of motor axons (Wang and Anderson, 1997; Helmbacher et al., 2000; Eberhart et al., 2004), commissural axons (Henkemeyer et al., 1996; Park et al., 1997), and mediate intraretinal axons (Birgbauer et al., 2000, 2001; Vidovic and Marotte, 2003) in the vertebrate systems. Studies from invertebrate systems support a role for EphR signaling in the development of primary olfactory nerve pathway in the moth (Kaneko and Nighorn, 2003) and the formation of topographic patterns of axonal connectivity in the Drosophila visual system (Dearborn et al., 2002) and the mushroom body (Boyle et al., 2006), possibly utilizing similar mechanisms.
On the basis of our observations, we suggest that AmEphR/ephrin systems play distinct stage dependent roles in the development of the honeybee brain. However, localization of EphR and ephrin proteins was examined using an in situ protein labeling technique with Fc-fusion probes. While labeling with Fc-fusion probes comes with certain advantages, such as high specificity and capability of detecting all interacting partners, it is nevertheless considered less sensitive than immunohistochemistry (Flanagan, 2000; Kaneko and Nighorn, 2003), thus less efficient for protein detection. A possibility remains therefore that the level of the two proteins expressed in older Kenyon cells and in the neuropiles of the mushroom body may not be sufficiently high for detection by in situ labeling with Fc-fusion proteins. Alternatively, the lack of the two proteins detected in some regions of the developing brains and ephrin protein in brain of adults may indicate an absence of protein expression through post-transcriptional regulation. The former is less likely considering the high levels and uniform expression of the mRNA especially during development. Invertebrate specific antibodies for EphR/ephrin proteins would be useful but are currently unavailable.
MsEphR-Fc exerts an inhibitory effect on behavioral performance in memory formation at 24 h after training in the honeybee. Presumably, the soluble MsEphR-Fc is binding to endogenous Amephrins and thus preventing the AmEphR activation. These expectations were based on the findings from studies in mammalian systems where infusion of soluble EphA5-Fc was shown to impair the induction (Gao et al., 1998) and the maintenance (Gerlai et al., 1999; Gerlai, 2001) of long term potentiation (LTP) in area CA1 of hippocampal slices and in vivo treatment with EphA5-Fc impaired behavioral performance in hippocampal-dependent memory tasks in mice (Gerlai, 2001). However, treatment with ephrin-A5-IgG in a form (clustered) that allowed it to mimic the behavior of membrane bound ephrin-A ligands, acted as an activator of EphA5 to enhance LTP (Gao et al., 1998; Gerlai et al., 1999; Gerlai, 2001) and learning behavior in mice (Gerlai et al., 1999). In our study, similar results on behavioral performance in memory formation at 24 h were obtained with Msephrin-Fc and MsEphR-Fc treated honeybees because we used unclustered forms of Msephrin-Fc. The resultant amnesic effect was observed from a blockade of AmEphRs by Msephrin-Fc, thus making them inaccessible to the endogenous ligand (antagonist).
Reduction in the retention of associative learning at 1 h post-training, in individuals treated with ephrin-Fc 1 h prior to or 1 h post-training, was also demonstrated. As ephrin-Fc treatment affects memory of the learnt task 1 h after training, which persisted for at least 24 h, suggests that the amnesic effect results from blockade of existing AmEphRs and could potentially affect acquisition and the subsequent consolidation and or recall of the memory process. The fact that our results were obtained with a single injection of either Fc protein suggests that these reagents are stable in vivo not only to block activation of already existing EphR or ligand proteins, but may also inhibit the activation of newly synthesized proteins necessary for long term memory formation.
The localization of AmEphR protein to regions of the neuropile involved in associative learning and memory such as the mushroom bodies and antennal lobes (Müller, 2000; Menzel, 2001) is consistent with their involvement in the processing of sensory inputs and high order brain functions. AmEphR shares sequence similarities with both the mammalian EphR-A and EphR-B subclass members and may represent a prototypical Eph family member, as suggested for D-Eph (Scully et al., 1999) and MsEphR (Kaneko and Nighorn, 2003). Thus, it is tempting to speculate that the invertebrate EphR participates in learning and memory by regulating both the structural and physiological properties of synapses. Candidate receptors in the honeybee to which the AmEphR might bind, to modulate function, include putative NMDA and mGluRs glutamate receptors. Genomic data point to highly conserved genes that encode both ionotropic and metabotropic glutamate receptors (Ultsch et al., 1992, 1993; Parmentier et al., 1996; Volkner et al., 2000; Funada et al., 2004). Studies have suggested that glutamate is involved in long term memory formation in the honeybee (Maleszka et al., 2000; Locatelli et al., 2005). These effects are most likely mediated through the activated NMDA receptors as was demonstrated with NMDA receptor antagonists in the honeybee (Si et al., 2003) and genetic tools in the Drosophila (Xia et al., 2005).
In summary, the expression of AmEphR and Amephrin genes in developing and adult brain suggests a role in early developmental events and function in adult plasticity. The latter is supported by the effects of disruption of Eph signaling on memory. This is the first demonstration that Eph molecules function to regulate the formation of memory in insects. This indicates that the functions of EphR/ephrin in adult plasticity are highly conserved across diverse phyla.
Footnotes
Published online in Wiley InterScience(www.interscience.wiley.com).
We are indebted to Paul Helliwell for rearing Apis mellifera and for carrying out the conditioning and testing procedures. We thank Dr. Lauren Marotte for critical review of this manuscript. We also thank Sharyn Wragg for her excellent contribution in the preparation of illustrations and Sylvain Foret for assistance with statistical analysis.
References
- Aoki M, Yamashita T, Tohyama M. EphA receptors direct the differentiation of mammalian neural precursor cells through a mitogen-activated protein kinase-dependent pathway. J Biol Chem. 2004;279:32643–32650. doi: 10.1074/jbc.M313247200. [DOI] [PubMed] [Google Scholar]
- Birgbauer E, Cowan CA, Sretavan DW, Henkemeyer M. Kinase independent function of EphB receptors in retinal axon pathfinding to the optic disc from dorsal but not ventral retina. Development. 2000;127:1231–1241. doi: 10.1242/dev.127.6.1231. [DOI] [PubMed] [Google Scholar]
- Birgbauer E, Oster SF, Severin CG, Sretavan DW. Retinal axon growth cones respond to EphB extracellular domains as inhibitory axon guidance cues. Development. 2001;128:3041–3048. doi: 10.1242/dev.128.15.3041. [DOI] [PubMed] [Google Scholar]
- Bitterman M, Menzel R, Fietz A, Schäfer S. Classical conditioning of proboscis extension in honeybees (Apis mellifera) J Comp Psychol. 1983;97:107–119. [PubMed] [Google Scholar]
- Bossing T, Brand A. Dephrin, a transmembrane ephrin with a unique structure, prevents interneuronal axons from exiting the Drosophila embryonic CNS. Development. 2002;129:4205–4218. doi: 10.1242/dev.129.18.4205. [DOI] [PubMed] [Google Scholar]
- Boyle M, Nighorn A, Thomas JB. Drosophila Eph receptor guides specific axon branches of mushroom body neurons. Development. 2006;133:1845–1854. doi: 10.1242/dev.02353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SM, Napper RM, Thompson CM, Mercer AR. Stereological analysis reveals striking differences in the structural plasticity of two readily identifiable glomeruli in the antennal lobes of the adult worker honeybee. J Neurosci. 2002;22:8514–8522. doi: 10.1523/JNEUROSCI.22-19-08514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchert M, Schneider S, Meskenaite V, Adams MT, Canaani E, Baechi T, Moelling K, et al. The junction-associated protein AF-6 interacts and clusters with specific Eph receptor tyrosine kinases at specialized sites of cell-cell contact in the brain. J Cell Biol. 1999;144:361–371. doi: 10.1083/jcb.144.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Rogers C, Maron C, Henkemeyer M, Swanson GT, Heinemann SF. Trans-synaptic Eph receptor-ephrin signaling in hippocampal mossy fiber LTP. Science. 2002;296:1864–1869. doi: 10.1126/science.1069081. [DOI] [PubMed] [Google Scholar]
- Dacher M, Lagarrigue A, Gauthier M. Antennal tactile learning in the honeybee: Effects of nicotinic antagonists on memory dynamics. Neuroscience. 2005;130:37–50. doi: 10.1016/j.neuroscience.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- Davis S, Gale NW, Aldrich TH, Maisonpierre PC, Lhotak V, Pawson T, Goldfarb M, et al. Ligands for EPH-related receptors that require membrane attachment or clustering for activity. Science. 1994;266:816–819. doi: 10.1126/science.7973638. [DOI] [PubMed] [Google Scholar]
- Dearborn R, Jr, He Q, Kunes S, Dai Y. Eph receptor tyrosine kinase-mediated formation of a topographic map in the Drosophila visual system. J Neurosci. 2002;22:1338–1349. doi: 10.1523/JNEUROSCI.22-04-01338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depaepe V, Suarez-Gonzalez N, Dufour A, Passante L, Gorski JA, Jones KR, Ledent C, et al. Ephrin signalling controls brain size by regulating apoptosis of neural progenitors. Nature. 2005;435:1244–1250. doi: 10.1038/nature03651. [DOI] [PubMed] [Google Scholar]
- Drescher U, Kremoser C, Handwerker C, Loschinger J, Noda M, Bonhoeffer F. In vitro guidance of retinal ganglion cell axons by RAGS, a 25 kDa tectal protein related to ligands for Eph receptor tyrosine kinases. Cell. 1995;82:359–370. doi: 10.1016/0092-8674(95)90425-5. [DOI] [PubMed] [Google Scholar]
- Eberhart J, Barr J, O’Connell S, Flagg A, Swartz ME, Cramer KS, Tosney KW, et al. Ephrin-A5 exerts positive or inhibitory effects on distinct subsets of EphA4-positive motor neurons. J Neurosci. 2004;24:1070–1078. doi: 10.1523/JNEUROSCI.4719-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmer B, Gronenberg W. Segregation of visual inputs to the mushroom bodies in the honeybee (Apis mellifera) J Comp Neurol. 2002;451:362–373. doi: 10.1002/cne.10355. [DOI] [PubMed] [Google Scholar]
- Ethell IM, Irie F, Kalo MS, Couchman JR, Pasquale EB, Yamaguchi Y. EphB/syndecan-2 signaling in dendritic spine morphogenesis. Neuron. 2001;31:1001–1013. doi: 10.1016/s0896-6273(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Faber ESL, Sedlak P, Vidovic M, Sah P. Synaptic activation of transient receptor potential channels by metabotropic glutamate receptors in the lateral amygdale. Neuroscience. 2005;137:781–794. doi: 10.1016/j.neuroscience.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Farooqui T, Robinson K, Vaessin H, Smith BH. Modulation of early olfactory processing by an octopaminergic reinforcement pathway in the honeybee. J Neurosci. 2003;23:5370–5380. doi: 10.1523/JNEUROSCI.23-12-05370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SM, Abrams AI, Strausfeld NJ. Development and morphology of class II Kenyon cells in the mushroom bodies of the honey bee, Apis mellifera. J Comp Neurol. 2004;474:325–339. doi: 10.1002/cne.20146. [DOI] [PubMed] [Google Scholar]
- Farris SM, Robinson GE, Davis RL, Fahrbach SE. Larval and pupal development of the mushroom bodies in the honey bee, Apis mellifera. J Comp Neurol. 1999;414:97–113. doi: 10.1002/(sici)1096-9861(19991108)414:1<97::aid-cne8>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Flanagan JG. In situ analysis of embryos with receptor or ligand fusion protein probes. Curr Biol. 2000;10:R52–R53. doi: 10.1016/s0960-9822(00)00293-1. [DOI] [PubMed] [Google Scholar]
- Funada M, Yasuo S, Yoshimura T, Ebihara S, Sasagawa H, Kitagawa Y, Kadowaki T. Characterization of the two distinct subtypes of metabotropic glutamate receptors from honeybee, Apis mellifera. Neurosci Lett. 2004;359:190–194. doi: 10.1016/j.neulet.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Ganeshina O, Schäfer S, Malun D. Proliferation and programmed cell death of neuronal precursors in the mushroom bodies of the honeybee. J Comp Neurol. 2000;417:349–365. [PubMed] [Google Scholar]
- Gao PP, Sun CH, Zhou XF, DiCicco-Bloom E, Zhou R. Ephrins stimulate or inhibit neurite outgrowth and survival as a function of neuronal cell type. J Neurosci Res. 2000;60:427–436. doi: 10.1002/(SICI)1097-4547(20000515)60:4<427::AID-JNR1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Gao WQ, Shinsky N, Armanini MP, Moran P, Zheng JL, Mendoza-Ramirez JL, Phillips HS, et al. Regulation of hippocampal synaptic plasticity by the tyrosine kinase receptor, REK7/EphA5, and its ligand, AL-1/Eph-rin-A5. Mol Cell Neurosci. 1998;11:247–259. doi: 10.1006/mcne.1998.0696. [DOI] [PubMed] [Google Scholar]
- Gascuel J, Masson C. Developmental study of afferented and deafferented bee antennal lobes. J Neurobiol. 1991;22:795–810. doi: 10.1002/neu.480220802. [DOI] [PubMed] [Google Scholar]
- Gerlai R. Eph receptors and neural plasticity. Nat Rev Neurosci. 2001;2:205–209. doi: 10.1038/35058582. [DOI] [PubMed] [Google Scholar]
- Gerlai R, Shinsky N, Shih A, Williams P, Winer J, Armanini M, Cairns B, et al. Regulation of learning by EphA receptors: A protein targeting study. J Neurosci. 1999;19:9538–9549. doi: 10.1523/JNEUROSCI.19-21-09538.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald C, Korte M, Wolfer D, Wilkinson GA, Unsicker K, Lipp HP, Bonhoeffer T, et al. Kinase-independent requirement of EphB2 receptors in hippocampal synaptic plasticity. Neuron. 2001;32:1027–1040. doi: 10.1016/s0896-6273(01)00550-5. [DOI] [PubMed] [Google Scholar]
- Hähnlein I, Bicker G. Morphology of neuroglia in the antennal lobes and mushroom bodies of the brain of the honeybee. J Comp Neurol. 1996;367:235–245. doi: 10.1002/(SICI)1096-9861(19960401)367:2<235::AID-CNE6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Helmbacher F, Schneider-Maunoury S, Topilko P, Tiret L, Charnay P. Targeting of the EphA4 tyrosine kinase receptor affects dorsal/ventral pathfinding of limb motor axons. Development. 2000;127:3313–3324. doi: 10.1242/dev.127.15.3313. [DOI] [PubMed] [Google Scholar]
- Henderson JT, Georgiou J, Jia ZP, Robertson J, Elowe S, Roder JC, Pawson T. The receptor tyrosine kinase EphB2 regulates NMDA-dependent synaptic function. Neuron. 2001;32:1041–1056. doi: 10.1016/s0896-6273(01)00553-0. [DOI] [PubMed] [Google Scholar]
- Henkemeyer M, Orioli D, Henderson JT, Saxton TM, Roder J, Pawson T, Klein R. Nuk controls path-finding of commissural axons in the mammalian central nervous system. Cell. 1996;86:35–46. doi: 10.1016/s0092-8674(00)80075-6. [DOI] [PubMed] [Google Scholar]
- Himanen JP, Nikolov DB. Eph receptors and ephrins. Int J Biochem Cell Biol. 2003;35:130–134. doi: 10.1016/s1357-2725(02)00096-1. [DOI] [PubMed] [Google Scholar]
- Holland SJ, Gale NW, Mbamulu G, Yancopoulos GD, Henkemeyer M, Pawson T. Bidirectional signaling through the Eph-family receptor Nuk and its transmembrane ligands. Nature. 1996;383:722–725. doi: 10.1038/383722a0. [DOI] [PubMed] [Google Scholar]
- Holmberg J, Armulik A, Senti K-A, Edoff K, Spalding K, Momma S, Cassidy R, et al. Ephrin-A2 reverse signaling negatively regulates neural progenitor proliferation and neurogenesis. Genes Dev. 2005;19:462–471. doi: 10.1101/gad.326905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Nighorn A. Interaxonal Eph-ephrin signaling may mediate sorting of olfactory sensory axons in Menduca sexta. J Neurosci. 2003;23:11523–11538. doi: 10.1523/JNEUROSCI.23-37-11523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull CE, Lansford R, Gale NW, Collazo A, Marcelle C, Yancopoulos GD, et al. Interactions of Fph related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Curr Biol. 1997;7:571–580. doi: 10.1016/s0960-9822(06)00256-9. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleszka R. Evaluation of differential gene expression during behavioral development in the honeybee using microarrays and northern blots. Genome Biol. 2002;3:7.1–7.9. doi: 10.1186/gb-2002-3-2-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R, Maleszka R. Transcriptional profiling reveals multifunctional roles for transferrin in the honeybee (Apis mellifera) J Insect Sci. 2003;3:27–36. doi: 10.1093/jis/3.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshan PT, Hamilton IS, Mustard JA, Mercer AR. Developmental changes in expression patterns of two do-pamine receptor genes in mushroom bodies of the honeybee, Apis mellifera. J Comp Neurol. 2003;466:91–103. doi: 10.1002/cne.10864. [DOI] [PubMed] [Google Scholar]
- Liebl DJ, Morris CJ, Henkemeyer M, Parada LF. mRNA expression of ephrins and Eph receptor tyrosine kinases in the neonatal and adult mouse central nervous system. J Neurosci Res. 2003;71:7–22. doi: 10.1002/jnr.10457. [DOI] [PubMed] [Google Scholar]
- Locatelli F, Bundrock G, Müller U. Focal and temporal release of glutamate in the mushroom bodies improves olfactory memory in Apis mellifera. J Neurosci. 2005;25:11614–11618. doi: 10.1523/JNEUROSCI.3180-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleszka R, Helliwell P. Effect of juvenile hormone on short-term olfactory memory in young honeybees Apis mellifera. Horm Behav. 2001;40:403–408. doi: 10.1006/hbeh.2001.1705. [DOI] [PubMed] [Google Scholar]
- Maleszka R, Helliwell P, Kucharski R. Pharmacological interference with glutamate re-uptake impairs long-term memory in the honeybee Apis mellifera. Behav Brain Res. 2000;115:49–53. doi: 10.1016/s0166-4328(00)00235-7. [DOI] [PubMed] [Google Scholar]
- Malun D. Early development of mushroom bodies in the brain of the honeybee Apis mellifera as revealed by BrdU incorporation and ablation experiments. Learn Mem. 1998;5:90–101. [PMC free article] [PubMed] [Google Scholar]
- Marotte LR, Vidovic M, Wheeler E, Jhaveri S. Brain-derived neurotrophic factor is expressed in a gradient in the superior colliculus during development of the retino-collicular projection. Eur J Neurosci. 2004;20:843–847. doi: 10.1111/j.1460-9568.2004.03521.x. [DOI] [PubMed] [Google Scholar]
- Martinez A, Soriano E. Functions of ephrin/Eph interactions in the development of the nervous system: Emphasis on the hippocampal system. Brain Res Rev. 2005;49:211–226. doi: 10.1016/j.brainresrev.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Martone ME, Holash JA, Bayardo A, Pasquale EB, Ellisman MH. Immunolocalization of the receptor tyro-sine kinase EphA4 in the adult rat central nervous system. Brain Res. 1997;771:238–250. doi: 10.1016/s0006-8993(97)00792-0. [DOI] [PubMed] [Google Scholar]
- Menzel R. Learning, memory, and ‘‘cognition’’ in honey bees. In: Kesner RP, Otten DS, editors. Neurobiology of Comparative Cognition. Hillsdale, NJ: Erlbaum; 1990. pp. 237–292. [Google Scholar]
- Menzel R. Searching for the memory trace in a mini-brain, the honeybee. Learn Mem. 2001;8:53–62. doi: 10.1101/lm.38801. [DOI] [PubMed] [Google Scholar]
- Mobbs PG. The brain of the honeybee Apis mellifera. I. The connections and spatial organization of the mushroom bodies. Philos Trans R Soc Lond B Biol Sci. 1982;298:309–354. [Google Scholar]
- Müller U. Prolonged activation of cAMP dependent protein kinase during conditioning induces long term memory in honeybees. Neuron. 2000;27:159–168. doi: 10.1016/s0896-6273(00)00017-9. [DOI] [PubMed] [Google Scholar]
- Murai KK, Nguyen LN, Irie F, Yamaguchi Y, Pasquale EB. Control of hippocampal dendritic spine morphology through ephrin-A3/EphA4 signaling. 2003;6:153–160. doi: 10.1038/nn994. [DOI] [PubMed] [Google Scholar]
- Murai KK, Pasquale EB. Eph receptors, ephrins and synaptic function. Neuroscientist. 2004;10:304–314. doi: 10.1177/1073858403262221. [DOI] [PubMed] [Google Scholar]
- Osborne PB, Vidovic M, Chieng B, Hill CE, Christie MJ. Expression of mRNA and functional α1-adreno-ceptors that suppress the GIRK conductance in adult rat locus coeruleus neurons. Br J Pharmacol. 2002;135:226–232. doi: 10.1038/sj.bjp.0704453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Frisen J, Barbacid M. Aberrant axonal projections in mice lacking EphA8 (Eek) tyrosine protein kinase receptors. EMBO J. 1997;16:3106–3114. doi: 10.1093/emboj/16.11.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier M, Pin J, Bockaert J, Grau Y. Cloning and functional expression of a Drosophila metabotropic glutamate receptor expressed in the embryonic CNS. J Neurosci. 1996;16:6687–6694. doi: 10.1523/JNEUROSCI.16-21-06687.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribi WA, Scheel M. The second and third optic ganglia of the worker bee. Cell Tissue Res. 1981;221:17–43. doi: 10.1007/BF00216567. [DOI] [PubMed] [Google Scholar]
- Scalia F, Feldheim DA. Eph/ephrin A-and B-family expression patterns in the leopard frog (Rana utricularia) Dev Brain Res. 2005;158:102–106. doi: 10.1016/j.devbrainres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Scully AL, McKeown M, Thomas JB. Isolation and characterization of Dek, a Drosophila Eph receptor protein tyrosine kinase. Mol Cell Neurosci. 1999;13:337–347. doi: 10.1006/mcne.1999.0752. [DOI] [PubMed] [Google Scholar]
- Si A, Helliwell P, Maleszka R. Effects of NMDA receptor antagonists on olfactory learning and memory in the honeybee (Apis mellifera) Pharmacol Biochem Behav. 2003;77:191–197. doi: 10.1016/j.pbb.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, et al. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Strausfeld NJ, Homberg U, Kloppenburg P. Parallel organization in honey bee mushroom bodies by peptidergic Kenyon cells. J Comp Neurol. 2000;424:179–195. [PubMed] [Google Scholar]
- Takasu MA, Dalva MB, Zigmond RE, Greenberg ME. Modulation of NMDA receptor-dependent calcium influx and gene expression through EphB receptors. Science. 2002;295:491–495. doi: 10.1126/science.1065983. [DOI] [PubMed] [Google Scholar]
- Torres R, Firestein BL, Dong H, Staudinger J, Olson EN, Huganir RL, et al. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- Ultsch A, Schuster CM, Laube B, Betz H, Schmitt B. Glutamate receptors of Drosophila melanogaster. Primary structure of a putative NMDA receptor protein expressed in the head of the adult fly. FEBS Lett. 1993;324:171–177. doi: 10.1016/0014-5793(93)81387-f. [DOI] [PubMed] [Google Scholar]
- Ultsch A, Schuster CM, Laube B, Schloss P, Schmitt B, Betz H. Glutamate receptors of Drosophila melanogaster: Cloning of a kainate-selective subunit expressed in the central nervous system. Proc Natl Acad Sci USA. 1992;89:10484–10488. doi: 10.1073/pnas.89.21.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic M, Marotte LR. Analysis of EphB receptors and their ligands in the developing retinocollicular system of the wallaby reveals dynamic patterns of expression in the retina. Eur J Neurosci. 2003;18:1549–1558. doi: 10.1046/j.1460-9568.2003.02882.x. [DOI] [PubMed] [Google Scholar]
- Vidovic M, Marotte LR, Mark RF. Marsupial retino-collicular system shows differential expression of messenger RNA encoding EphA receptors and their ligands during development. J Neurosci Res. 1999;57:244–254. doi: 10.1002/(SICI)1097-4547(19990715)57:2<244::AID-JNR10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Volkner M, Lenz-Bohme B, Betz H, Schmitt B. Novel CNS glutamate receptor subunit genes of Drosophila melanogaster. J Neurochem. 2000;75:1791–1799. doi: 10.1046/j.1471-4159.2000.0751791.x. [DOI] [PubMed] [Google Scholar]
- Wang HU, Anderson DJ. Eph family transmembrane ligands can mediate repulsive guidance of the trunk neural crest migration and motor axon outgrowth. Neuron. 1997;18:383–396. doi: 10.1016/s0896-6273(00)81240-4. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. Eph receptors and ephrins: Regulators of guidance and assembly. Int Rev Cytol. 2000;196:177–244. doi: 10.1016/s0074-7696(00)96005-4. [DOI] [PubMed] [Google Scholar]
- Winslow JW, Moran P, Valverde J, Shih A, Yuan JQ, Wong SC, et al. Cloning of AL-1, a ligand for an Eph-related tyrosine kinase receptor involved in axon bundle formation. Neuron. 1995;14:973–981. doi: 10.1016/0896-6273(95)90335-6. [DOI] [PubMed] [Google Scholar]
- Xia S, Miyashita T, Fu TF, Lin WY, Wu CL, Pyzocha L, Lin IR, et al. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr Biol. 2005;15:603–615. doi: 10.1016/j.cub.2005.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Alldus G, Holder N, Wilkinson DG. Expression of truncated Sek-1 receptor tyrosine kinase disrupts the segmental restriction of gene expression in the Xenopus and zebrafish hindbrain. Development. 1995;121:4005–4016. doi: 10.1242/dev.121.12.4005. [DOI] [PubMed] [Google Scholar]
- Zhou X, Suh J, Cerretti DP, Zhou R, DiCicco-Bloom E. Ephrins stimulate neurite outgrowth during early cortical neurogenesis. J Neurosci Res. 2001;15:1054–1063. doi: 10.1002/jnr.10029. [DOI] [PubMed] [Google Scholar]


