Abstract
This paper describes a frame-based integration of the three GO subontologies, the Chemicals of Biological Interest ontology (ChEBI), and the Cell Type Ontology (CTO) in which relationships between elements of the ontologies are modeled in a way that better captures the relational semantics between biological concepts represented by the terms, rather than between the terms themselves, than previous frame-based efforts. We also describe a methodology for creating suggested enriching assertions of the form (subject, relationship, object) by identifying patterns in GO terms, mapping these patterns and subpatterns to relationships, matching concepts to these patterns and subpatterns, and integrating these assertions into the ontologies. Using this methodology, a large number of reliable assertions linking previously unlinked OBO terms using a wide variety of specific, hierarchically arranged relationships were created: A predicted assertion was made for 62% of GO terms that matched one of 31 patterns, and 97% of these predicted assertions were assessed to be valid; a further 429 assertions (corresponding to 6% of the matching terms) were manually created, resulting in an initial set of 4,497 assertions. Furthermore, this methodology programmatically integrates assertions into a base ontology such that each assertion is fully consistent with respect to higher (i.e., more general) relevant class and slot levels. Such an integration is absent from previous compositional efforts, and we argue its necessity for the creation of coherent biological ontologies when linking previously unlinked terms.
1. Introduction
The work presented in this paper demonstrates that OBO ontologies can be reliably semantically enriched through the creation of a large number of associations between existing biological concepts using specific relationships and careful integration of these associations into a consistent, unified ontology. An enormous effort has gone into the creation and maintenance of the OBO ontologies [1], and this has been met with their ever-increasing use by biological researchers. There are currently over 50 OBO ontologies, ranging over such domains as anatomy, behavior, phenotype, experiment, and sequence. The flagship OBO ontology, the Gene Ontology (GO) [2], with its three subontologies detailing molecular functions, biological processes, and cellular components, in particular has experienced a phenomenal growth in terms of numbers of concepts and also of its extensive use by annotators to describe gene and gene-product entries in a number of prominent model-organism databases.
Although many of these ontologies are large, they are structurally quite simple, typically consisting of only a few relationship types apart from the fundamental is-a relationship that forms the backbone of an ontology. Furthermore, there are no links between terms from separate ontologies, even between terms from different GO subontologies. To be fair, the OBO ontologies were designed to eschew such linkages, with the rationale that the terms could be applied to genes and gene-product entries in biological databases without additional, possibly erroneous implications. However, most concepts do have relationships with other concepts, as evidenced in their natural-language names and definitions, and computational agents cannot take advantage of these relationships if they are not formally represented. The compositionality of GO terms has been noted [3, 4], and there have been efforts, most notably the GONG [5, 6] and Obol [7] projects, that have taken advantage of this compositionality to produce more formal definitions of GO terms in the form of added relationships with other ontological terms. The work presented here expands on these efforts by using a hierarchical set of specific relationships and OBO terms to create a large number of assertions that are consistently integrated into the component ontologies to create a much more richly connected network of biological concepts.
This paper describes a frame-based integration of the three GO subontologies, the Chemicals of Biological Interest ontology (ChEBI) [8], and the Cell Type Ontology (CTO) [9] in which relationships between elements of the ontologies are modeled in a way that better captures the relational semantics between biological concepts represented by the terms, rather than between the terms themselves, than previous frame-based efforts. We also describe a methodology for creating suggested enriching assertions of the form (subject, relationship, object) by identifying patterns in GO terms, mapping these patterns and subpatterns to relationships, matching concepts to these patterns and subpatterns, and integrating these assertions into the ontologies. Using this methodology, a large number of reliable assertions linking previously unlinked OBO terms using a wide variety of specific, hierarchically arranged relationships were created: A predicted assertion was made for 62% of GO terms that matched one of 31 patterns, and 97% of these predicted assertions were assessed to be valid; a further 429 assertions (corresponding to 6% of the matching terms) were manually created, resulting in an initial set of 4,497 assertions. Furthermore, this methodology programmatically integrates assertions into a base ontology such that each assertion is fully consistent with respect to higher (i.e., more general) relevant class and slot levels. Such an integration is absent from previous compositional efforts, and we argue its necessity for the creation of coherent biological ontologies when linking previously unlinked terms.
2. Methods and Rationale
2.1 Representation of GO, ChEBI, and CTO
The ontologies used in this project were obtained from the OBO Web site as OBO files. The three subontologies of GO were included in this enrichment effort because they are the most widely used of the OBO ontologies in biological research. In addition to these, ChEBI and CTO were selected because (a) they are sufficiently advanced; (b) they were expected to suggest many types of relationships and many assertions using these relationships between the terms of these two ontologies and those of the GO subontologies; and (c) these ontologies are largely orthogonal, thus avoiding complicated ontology-merging issues. A frame-based representation was used for the integrated ontology because it is a relatively intuitive, widely used knowledge model that also comes with a sophisticated, powerful set of tools (particularly Protege [10] and its many associated applications) that can be used to create, edit, and programmatically manipulate complex ontologies. This decision was also influenced by the intention to eventually use the resulting integrated ontology in natural-language-processing tasks in the biomedical domain [11].
The Protege OBO Import Tab [12] was used to create initial frame-based versions of the GO subontologies, ChEBI, and CTO. This frame-based representation of OBO ontologies is similar to that of Yeh et al. [13] in that each term is assigned a class, and the term attributes (e.g., name, synonyms, definition, definition reference) are stored in the respective term’s frame as values of slots defined for a metaclass of OBO or GO terms. This information is properly modeled as metadata about the terms themselves. However, both Yeh et al. and the Protege OBO Import Tab also define relationships (e.g., part_of in GO, develops_from in CTO) at the metaclass level (i.e., a metaclass of the OBO terms is made the domain of the slot), so these relationships also appear as term metadata. This approach models relationships as properties linking terms, e.g., that the term nucleus is part of the term cell, rather than a relationship between modeled concepts, e.g., that a nucleus can be part of a cell. Also, when modeled as metaclass data, this relational information is not inherited either by instances or by subclasses. For example, in a frame-based GO cellular-component ontology created by the OBO Import Tab, a created instance of nucleus (which represents a specific real or simulated nucleus) does not inherit its partitive relationship to a cell, nor does any of its subclasses (e.g., macronucleus). Thus, an application processing an instance of nucleus could not infer that it can be part of a cell without additionally processing metaclass information from its parent class. Furthermore, in a representation in which relationships are defined at the level of a metaclass, each relationship appears in every ontology class that is an instance of this metaclass. Using such a representation, the relationship results in formation of, for example, would also appear as a slot for each term, the vast majority of which are not appropriate as the domain of this relationship.
For these reasons, relationships were changed from the metaclass-level representation produced by the OBO Import Tab to a representation at the class level. That is, the domains of all relationships were set to the term classes themselves rather than a metaclass of terms. Thus, in our representation, part_of is attached to the class nucleus (among others), and cell is its corresponding allowed class. This representation more precisely captures the semantics, in this case, that a nucleus can be a part of a cell, as opposed to the term nucleus being a part of the term cell. This partitive relationship is inherited by any created instance of the class nucleus (i.e., any direct instance of nucleus or an instance of one of its subclasses) such that the instance can be asserted to be part of an instance of a cell. Thus, an instance’s relationships to other instances is represented within the frame of the instance and can be processed by an agent without having to also process the frames of the class(es) to which the instance belongs. In addition, in this representation, a given relationship is attached only to those concepts for which the relationship is appropriate. For example, results in formation of is applied only to those GO terms (e.g., ketone biosynthesis and its subclasses) for which it has been asserted that an entity is being formed.
Some of the OBO ontologies, including GO and CTO, suffer from the problem of having orphan terms—terms that have no parents via the is-a relationship. Both Yet et al. and the Protege OBO Import Tab capture orphan terms in special classes, an approach that was also taken in our effort. Any orphan term can by default be made a direct subclass of : THING, Protege’s system root class, so this step was not done out of necessity but rather for usability purposes: Instead of having a long list of these terms placed directly under : THING, they are now neatly integrated under their corresponding part-of-subontology concepts; such a representation is convenient when making a query of a particular subontology. An OBO term metaclass was also created, with GO term, ChEBI term, and CTO term as submetaclasses. Every OBO term is an instance of the submetaclass corresponding to the ontology to which it belongs, while the classes created by us described above are instances only of the system class : STANDARD-CLASS. This allows accurate retrieval and navigation of a particular OBO-term hierarchy in the integrated system.
2.2 Finding Patterns in OBO Terms
This proposed methodology for creating enriching assertions between OBO terms is based on finding patterns in GO terms. Here, a pattern refers to a regular expression to which a string with an invariant substring and a variable substring of one or more characters can be matched. GO terms (both primary name and their synonyms) are the strings that are examined, and either an entire GO term or a substring of the term can match the pattern. In addition, the text that matches the variable part of the pattern must be able to be mapped to another OBO term (either a primary name or a synonym, analogously). A term that contains an embedded term generally implies that there is some sort of relationship, direct or indirect, between the two terms. Therefore, such patterns enable the generation of assertions consisting of relationships between terms.
For example, ( [^\n]+)\smembrane is a pattern, using Java regular-expression syntax, consisting of one or more non-new-line characters (i.e., any character of a GO term), followed by a single whitespace character, followed by the string “membrane”. The grouping, indicated by the parentheses, indicates that the substring that matches the enclosed subpattern [^\n]+ is the variable part that must be able to be mapped to another OBO term. Thus, this pattern can be applied to any GO term that in whole or in part consists of a substring of one or more characters that can be mapped to an OBO term, followed by one whitespace character and then the substring “membrane” (e.g., cell projection membrane, which contains a reference to the cellular-component term cell projection; or hydrogenosomal membrane, whose synonym hydrogenosome membrane contains a reference to the cellular-component term hydrogenosome).
To facilitate the mapping of text to concepts, all primary terms and their synonyms were canonicalized by removing each terminal “s” (e.g., to match the first two tokens of the GO biological-process term fatty acid oxidation to the ChEBI term fatty acids), converting each uppercase character to lowercase (e.g., to match the first token of the GO biological-process term acriflavine transport to the ChEBI term Acriflavine), and replacing each “-”, “\”, “(“, “)”, “_”, and “,” with a space (e.g., to match the first three tokens of the GO biological-process term natural killer cell proliferation to the CTO term natural_killer_cell). Patterns were searched for by tokenizing each term using a space as a delimiter (since other likely delimiters, listed above, have now been replaced with spaces) and then attempting to find concept matches in their canonical forms to maximum-length subsequences of the tokens. For example, for the GO biological-process term amino acid fermentation, a match was made using the first two tokens of the term ( amino acid) to the ChEBI term amino acids. If no matches were found, then matches of progressively shorter subsequences of term tokens to OBO terms were attempted. For example, for the GO biological-process term anaerobic gallate catabolism, after no match of length two was made, a match was made using the the middle token ( gallate) to the ChEBI term gallate. With this method, a match to the longest embedded OBO term is attempted, as this is the term that is most likely in a direct relationship with the base GO term.
This analysis of GO terms produced a list of patterns, ranked by frequency, each of which consists of an invariant part and a variable part to which other GO, ChEBI, or CTO terms match. The most frequently occurring patterns that also implied direct, reasonably semantically consistent relationships between the embedding and embedded OBO terms and that also could be fairly straightforwardly modeled in the ontology were manually selected for further processing. Furthermore, since the assertions that are to be created using these patterns are those in which terms that match the entire pattern are subjects of the assertions and terms that match a subpattern of the pattern (i.e., the variable part of the pattern) are objects of the assertions, patterns selected were those that could be used to create such assertions. In some cases, patterns that did not occur frequently but were natural complements to those that did (e.g., a pattern to match dephosphorylation terms to complement the pattern for the more frequently occurring phosphorylation terms) were added to this list.
There is not necessarily a simple one-to-one pattern-to-slot relationship: A given pattern generally does not imply the same relationship in all terms that match the pattern. For example, although the pattern ([^\n]+)\sbiosynthesis usually implies the assertion that the term matching the pattern results in the formation of the term matching the embedded subpattern, the GO biological-process term organismal biosynthesis is not the formation of an organism but the “transformation of simpler substances into more complex ones in multicellular organisms, occurring at the tissue, organ or organismal level”. On the other hand, a given relationship may not be implied by only one pattern. For example, both ([^\n]+)\scatabolism and ([^\n]+)\sdisassembly were mapped to the slot results in breakdown of in this study. A sufficient number of pattern were selected to create a list of 25 unique slots, from which 25 inverse slots were easily formed.
In Protege, slots may be arranged in hierarchies analogously to classes, an expressivity which is useful here. The advantage of such an organization is analogous to that of the arrangement of classes into a hierarchy: Just as asserting an entity to be a member of class implies that the entity is also a member of each of the class’s superclasses, making an assertion using a slot implies a set of assertions using each of the slot’s superslots. For example, results in oxidation of was made a direct subslot of results in modification of; the implication here is that if a process oxidizes a substance, it also necessarily modifies it. Thus, if an agent queries for processes in which a given entity has been modified, this hierarchical arrangement of slots can be used to also return processes in which the entity has been oxidized. The set of relationships and their hierarchical placement created for this project is not claimed to be optimal, but it is an expressive and useful representation of enriching relationships between OBO terms and is proposed as an initial basis for a community-consensus-driven representation.
For each of the chosen patterns, a list of all GO terms that match the pattern, in whole or in part, was created. The patterns were manually adjusted to appropriately expand or restrict the set of matching terms. For example, the pattern ([^\n]+)\smodification (for modification terms in which the text matching the grouped subpattern may refer to the entity that is being modified) was modified to (?!(?:positive\s|negative\s)*regulation\sof\s) ([^\n]+)\smodification[^\n]*. Here, the prefix was added so as to not capture modification terms that begin with “regulation of”, “positive regulation of”, or “negative regulation of” (e.g., regulation of histone modification), since the implied relationship between a term that matches such a pattern and the longest embedded term is not one of modification but rather one of regulation between the term and a modification process. The suffix was added so as to capture modification terms that have additional characters after “modification” (e.g., cell wall modification during abscission), for which the same modifying relationship may be implied.
2.3 Using Patterns to Create Assertions
For each term matching a pattern, the text matching the variable subpattern was mapped to an OBO term by comparing the matching string to canonicalized versions of primary term names and their synonyms. For a given pattern, this search was restricted to a subset of the ontologies representing a semantically appropriate range for the value of the relationship. For example, only the ChEBI and GO cellular-component ontologies were searched for the concepts referring to the entities being modified in GO entity-modification terms. Each mapping produces an ordered triple consisting of a subject OBO term (represented as a class), a relationship (represented as a slot that is attached to the subject class), and an object OBO term (represented as a class that is an allowed class of the slot) that is related to the subject term by the relationship, which we call an assertion. This assertional model is quite similar to the RDF triple, which consists of a subject, predicate, and object. Thus, each match resulted in a predicted assertion consisting of the original matching term (i.e., its class name), the implied relationship (i.e., a slot name), and the term to which the embedded term was matched (i.e., its class name), which was written to a text file.
As noted earlier, the fact that a given term matches a given pattern does not necessarily imply the relationship between the term and the embedded term to which the pattern was mapped. On the other hand, this relationship may hold, but the embedded text may be matched to the wrong OBO term. These are considered two distinct kinds of false positives. Thus, each predicted assertion was manually validated before being further processed. A domain expert checked each assertion by comparing it with the terms’ natural-language definitions, if present. In evaluating the assertions, it was considered not whether the predicted assertion must take place for a given instance of the subject term but rather if it could occur. This is justified by Protege’s knowledge model, in which the addition of a slot (along with its set of allowed classes) to a class does not imply that it holds for every created instance of the class (unless the slot is designated as required, which has not been done for any slot in this project as of yet). Rather, for a created instance of the class, if a value is entered for an added slot, it must be an instance of at least one of the slot’s allowed classes. (A default nonrequired slot is thus analogous to a universal restriction of a description logic.)
Manual validation also verified that the suggested object term matched the concept (or at least one of the concepts, none of which subsumes another) mentioned in the definition and not a subclass of it. For example, the hypothetical assertion [epithelial cell differentiation, results in differentiation into, duct_epithelial_cell] passes the criterion of possible occurrence, as an epithelial cell differentiation could result in the differentiation of a cell into a duct epithelial cell. However, the definition of epithelial cell differentiation is the set of processes “whereby a relatively unspecialized cell acquires specialized features of an epithelial cell, any of the cells making up an epithelium”. Considering this definition, the CTO term epithelial_cell, which is a superclass of duct_epithelial_cell, is the correct object term for this assertion. Thus, the hypothetical assertion with duct_epithelial_cell as the object term would have been assessed as incorrect. It was analogously required that the object term should not be a superclass of the object term (or one of the object terms) mentioned in the definition. For example, the hypothetical assertion [neuroblast proliferation (sensu Vertebrata), results in proliferation of, neuroblast] also passes the criterion of possible occurrence. However, neuroblast (sensu Vertebrata) (a subclass of neuroblast) is a better match as the object term, so the assertion with neuroblast as the object term would be judged incorrect.
During the evaluation of the predicted assertions, for each term matching a given pattern for which an assertion had not been predicted using the relationship to which the pattern had been mapped, a manual creation of an assertion through examination of the term, any embedded terms, and their definitions was attempted. For example, for the pattern (in a simplified format, as in Table 1) x lumen*, no assertion was predicted for the matching term vacuolar lumen using is lumen of, the relationship to which this pattern was mapped (as seen in Table 1), since the application could not map vacuolar to any OBO term. Manually finding the term vacuole in the GO cellular-component ontology, the assertion [vacuolar lumen, is lumen of, vacuole], stating that a vacuolar lumen can be asserted to be the lumen of a vacuole, was added. It is not claimed that this procedure catches all such false negatives. Rather, this step presents an opportunity to the evaluating expert to create an assertion for a given term and relationship that was not programmatically suggested.
Table 1.
Patterns used and the slots to which they have been mapped. The patterns were actually represented using Java regular-expression syntax in the project, but they are shown here in a simplified syntax for readability. x refers to one or more characters, and * refers to zero or more characters. Patterns used within the GO biological-process ontology were preceded by a negating element to avoid capturing terms beginning with “regulation of”, “negative regulation of”, or “positive regulation of”. Patterns have been mapped to the slots such that a term matching a pattern may be related to the term that matches the subpattern x via the specific slot.
| Patterns | Mapped Slot |
|---|---|
| x lumen* | is lumen of |
| x membrane* | is membrane of |
| x binding*
x docking* |
results in binding of |
| x catabolism*
x disassembly* |
results in breakdown of |
| x dephosphorylation* | results in dephosphorylation of |
| detection of x | results in detection of stimulus from |
| x development* | results in development of |
| x differentiation* | results in differentiation into |
| x inheritance* | results in distribution into daughter cells of |
| x division*
x fission* |
results in division of |
| x fermentation* | results in fermentation of |
| x assembly*
x biosynthesis* x formation* x salvage* |
results in formation of |
| x fusion* | results in fusion of |
| x maturation* | results in maturation into |
| x metabolism* | results in metabolism of |
| x modification* | results in modification of |
| negative regulation of x | results in negative regulation of |
| x oxidation* | results in oxidation of |
| x phosphorylation* | results in phosphorylation of |
| positive regulation of x | results in positive regulation of |
| x proliferation* | results in proliferation of |
| x reduction* | results in reduction of |
| regulation of x | results in regulation of |
| x secretion* | results in secretion of |
| x transport* | results in transport of |
For each validated assertion, an inverse assertion (using the inverse slot of the slot used in the original assertion) was also created. For example, the assertion [spectrin binding, results in binding of, spectrin] also results in the creation of the assertion [spectrin, binds in, spectrin binding]. As before, this does not state that a given molecule of spectrin must participate in a spectrin-binding event. Rather, if a molecule of spectrin binds to something, then it is participating in a spectrin-binding event.
It is important to note that although patterns within GO terms were extensively used, particularly for linking terms with terms found within these terms to form assertions, the methodology for integrating these assertions (described in later sections) does not depend on assertions that have such a relationship between the subject and object terms; any assertional triple consisting of a subject class, slot, and object class can be incorporated. In fact, some of the assertions manually added to the files during evaluation (for example, [guanine nucleotide transport, results in transport of, GTP], which asserts that a guanine-nucleotide transport event can result in the transport of a molecule of GTP) link subject terms with object terms that are not substrings of the subject terms. Patterns in GO terms were simply used to generate an initial list of straightforward relationships and a large initial set of assertions.
2.4 Primary Representation of Enriching Assertions
In a frame-based approach, assertions should be represented as slots and corresponding sets of allowed classes attached to classes, as slots linking classes are a central component of the frame-based knowledge model. However, in this representation, an allowed class that is subsumed by or equal to a class that has already been asserted as an allowed class of a given slot at a given level is not stored in order to avoid redundancy, as Protege interprets the setting of a slot’s allowed classes to two or more classes as a union of the classes. (For example, it is redundant to set the allowed classes of a slot to the set [binding, nucleotide binding], since this is equivalent to binding.) This is not a problem so long as the knowledge base is monotonic (i.e., assertions are never removed). However, the addition of an erroneous assertion to an ontology at some time and its later removal is a possibility. In some cases after removing an assertion, it would be necessary to set the allowed classes of the slot in question to a class stated in another assertion but one that is not stored in the final representation for the purpose of avoiding redundancy, as above. For example, if it were asserted that the value of a slot could be binding and, separately, nucleotide binding, only binding would be stored directly as an allowed class in this representation; if this assertion were later retracted, the slot should be set to nucleotide binding, but this could not be done since nucleotide binding was not stored directly as an allowed class. Thus, rather than storing our assertions only as slots with corresponding sets of allowed classes attached to classes, a primary representation to store all assertions assessed as reliable was used. With this representation, a consistent ontology can be recreated (as described in the next section) without losing knowledge if one or more assertions are later removed. The assertions were stored within the framework of the ontology itself, although, as we discuss later in this section, this particular method of storage is not the only possible choice.
Before describing in detail the knowledge model for the primary storage of assertions, it is useful to present the simpler underlying message: With this representation, a complete store of assertions that is used to set the allowed classes of the slots of the classes of the ontology (which is discussed in the next section) is maintained. For any given class, the other classes to which it is related (through the assertions) and the slots by which these classes are related are represented as instances that are directly linked to the class’s frame. This is convenient in that the set of assertions in which a given class is the subject is easily retrieved. However, it is necessary to iterate over these assertions if we wish to determine if a given class is the object of an assertion or even if a given class has at least one assertion via a given relationship. This is an inconvenience rather than a problem, as the Protege API can be used to answer such queries. Furthermore, classes do not have such large numbers of assertions that iteration over these instances to answer these types of queries has a significant performance penalty. Though this representation was used for the project thus far, this store could be implemented in other ways. For a very large store of assertions, a database would likely be more efficient than their representation within the ontology classes, as was done here. Nonetheless, this primary representation is sufficient for the work reported here.
Figure 1 displays a schema of the part of the ontology that was created to represent these manually validated assertions. The central class for the representation of assertions is the somewhat awkwardly but specifically named slot with direct allowed classes, which consists of a slot (which is the value of this class’s has slot slot) and a set of classes (which are the values of this class’s has direct allowed classes slot). The slot has slots with direct allowed classes is attached to the class OBO term, so this slot appears in each OBO term instance, and its values can be one or more instances of slot with direct allowed classes. The relationship and object OBO term of a given assertion are stored in an instance of slot with direct allowed classes (as values of has slot and has direct allowed classes, respectively), while the the subject OBO term of the assertion is the class to which this instance is linked via has slots with direct allowed classes. Each of these relationships has an inverse slot so that the links can be traversed bidirectionally.
Figure 1.

Knowledge model for the primary storage of assertions within the ontology. The relationship of a given assertion is made the value of has slot of an instance of slot with direct allowed classes, while the object term of the assertion is made a value of has direct allowed classes of the instance. This instance is made a value of has slots with direct allowed classes for the OBO term that is the subject term of the assertion. Each slot has an inverse slot so that the link between the two classes can be traversed bidirectionally.
As an example of this representation, consider ( cation transport, results in transport of, cations), i.e., the assertion that links the GO term cation transport to the ChEBI term z cations via the relationship results in transport of. First, an instance of slot with direct allowed classes is created. Within this instance, the slot results in transport of is made the value of the slot has slot, and the class cations is made a value of the slot has direct allowed classes; thus, this instance contains the second and third components of the assertion. This instance is then made a value of the slot has slots with direct allowed classes within the frame of the class cation transport.
The assertions were added one at a time to the ontology. For each assertion, if the subject term did not already have an attached instance of slot with direct allowed classes with the assertion’s relationship as the value of has slot, an instance was created, with the relationship the value of has slot and the object term a value of has direct allowed classes. If the subject term already had an attached instance of slot with direct allowed classes with the assertion’s relationship as the value of has slot, the object term was added as a value of has direct allowed classes to this existing instance.
2.5 Integration of Assertions into the Ontology
Once all valid assertions were stored in this primary representation, they were systematically integrated into the ontology. This integration, described below, creates slots attached to classes and corresponding sets of allowed classes such that the assertions are consistent with all assertions made at more general class and slot levels and such that there is no redundancy within the sets of allowed classes of slots. Assertions are processed one at a time, and the resulting ontology is independent of the order of the assertions.
In order to link previously unlinked classes of an ontology with a slot, the slot must be defined such that the domain and top-level set of allowed classes include the classes that are to be linked, either directly or indirectly through the inclusion of subsuming classes. More specifically, the domain must include the subject class to be linked, and the set of allowed classes must include the object class to be linked. If an expansion of one or both of these is necessary, the expansion should be minimal and should not include redundancy such that one of the classes subsumes another class within the set. Thus, it was first necessary to check, for each assertion, if the domain and/or top-level set of allowed classes of the slot of the assertion had to be expanded to accommodate the assertion. If the domain already contained the subject class of the assertion or contained a class that subsumes the subject class of the assertion, no expansion was needed; the top-level set of allowed classes was analogously checked to contain the object class of the assertion or a class that subsumes the object class of the assertion. If expansion was needed, it was checked that no remaining classes are subsumed by the newly added class; any that were found were removed from the set of classes in question to avoid redundancy.
In Protege, for a given slot, a top-level set of allowed classes is assigned. This top-level set may be narrowed at the level of a subclass to a subset of the top-level allowed classes. So, for a given assertion, since the object class of the assertion was integrated into the domain and top-level set of allowed classes of the slot of the assertion in the previous step, the object class was integrated at the level of the subject class. This was accomplished by checking to see if the set of allowed classes of the slot at the level of the subject class had to be analogously expanded to accommodate the object class of the assertion. If any expansion was needed, any redundant classes subsumed by the newly added class were removed, as before.
A class’s overriding set of allowed classes must also be a subset of the set of allowed classes of each of the class’s superclasses that is within the domain of a given slot. (Not all of the class’s superclasses will necessarily be within the domain of the slot.) Consider Figure 2a, which presents a simple example. Here, the GO term ammonia metabolism has been asserted to be related to the ChEBI term ammonia via the relationship results in metabolism of; that is, if this slot is given a value for an instance of ammonia metabolism, it must be an instance of ammonia. ammonia metabolism is a child of nitrogen compound metabolism, which is related to the ChEBI term nitrogen compounds via results in metabolism of. Since nitrogen compound metabolism is also within the domain of results in metabolism of, it must be checked that nitrogen compounds subsumes ammonia to avoid an inconsistency as described above. The example in Figure 2a actually poses no problem, as nitrogen compounds subsumes ammonia in ChEBI. Thus, this model is consistent in that ammonia metabolism can only result in the metabolism of ammonia, which is a nitrogen compound, and in that nitrogen compound metabolism, of which ammonia metabolism is a type, can only result in the metabolism of a nitrogen compound. Since these statements are consistent, nothing further needs to be done.
Figure 2.

Checking to propagate an allowed class up a class hierarchy. (a) There is no inconsistency between the assertions that nitrogen compound metabolism can only result in the metabolism of a nitrogen compound and that ammonia metabolism (which is a kind of nitrogen compound metabolism) can only result in the metabolism of ammonia, as ammonia is also a kind of nitrogen compound; therefore, nothing needs to be done. (b) There is an inconsistency between the assertions that nitrogen compound metabolism can only result in the metabolism of a nitrogen compound and that alkaloid metabolism (which is a kind of nitrogen compound metabolism) can only result in the metabolism of an alkaloid, as an alkaloid is not a kind of nitrogen compound. (c) To avoid this inconsistency, alkaloids is added to the set of allowed classes of results in metabolism of at the level of nitrogen compound metabolism.
The example in Figure 2b is problematic, however: The GO term alkaloid metabolism, another child of nitrogen compound metabolism, is asserted to be related to the ChEBI term alkaloids via results in metabolism of. The problem here stems from the fact that alkaloids is not subsumed by nitrogen compounds in ChEBI, while alkaloid metabolism is subsumed by nitrogen compound metabolism in GO. Thus, in this model, nitrogen compound metabolism can only result in the metabolism of a nitrogen compound, but alkaloid metabolism, which is a kind of nitrogen compound metabolism, can only result in the metabolism of an alkaloid, which is not a nitrogen compound. Our solution here is to integrate alkaloids into the set of allowed classes of results in metabolism of at the level of nitrogen compound metabolism (which just entails adding alkaloids as an allowed class, in this case), as seen in Figure 2c. In this model, nitrogen compound metabolism can only result in the metabolism of a nitrogen compound or an alkaloid, and alkaloid metabolism narrows this to an alkaloid. The potential inconsistency has been avoided.
Thus, it is necessary to check to propagate the assertion (using the slot and object class) up the class hierarchy so as not to introduce inconsistencies. That is, after the assertion was integrated at the level of the subject class assertion, the set of allowed classes of the slot at the level of each superclass within the domain of the slot was checked to see if it had to also be expanded to accommodate the object class of the assertion. Again, any redundant classes were removed at a given superclass level in the case of expansion. This process was performed recursively along each path within the domain of the given slot until it was not necessary for a set of allowed classes to be expanded.
Just as the set of allowed classes of a given slot at a given class level must be a subset of the set of allowed classes of the slot at the level of the class’s superclasses and at the top level, a set of allowed classes of a given slot must also be a subset of the set of allowed classes of the slot’s superslots. More simply, just as the constraints of a class should be more specific than those of its superclasses, the constraints of a slot should be more specific than those of its superslots. Consider Figure 3a, in which the ChEBI term lipids is asserted to be related to the GO term lipid modification via is modified in and to the GO term lipid phosphorylation via is phosphorylated in; furthermore, is phosphorylated in is a subslot of is modified in. It must be checked that lipid modification is a superclass of lipid phosphorylation. In this case, there is no inconsistency, as lipid phosphorylation is a subclass of lipid modification in GO.
Figure 3.

Checking to propagate an allowed class up a slot hierarchy. (a) There is no inconsistency between the assertions that a lipid can only be modified in a lipid-modification event and that a lipid can only be phosphorylated in a lipid-phosphorylation event, as a lipid phosphorylation is a kind of lipid modification, according to GO; thus, nothing further needs to be done in this case. (b) There is an inconsistency here in that an organic acid cannot be modified (since there is no explicit modification assertion), yet it can be phosphorylated, and thus modified (since is phosphorylated in is a subslot of is modified in), in an organic-acid-phosphorylation event. (c) To avoid this inconsistency, is modified in is applied to organic acids, and organic acid phosphorylation is made its allowed class. Now, an organic acid can be modified (or, more specifically, phosphorylated) in an organic-acid-phosphorylation event.
However, there is a problem in Figure 3b: The ChEBI term organic acids is asserted to be related to the GO term organic acid phosphorylation via is phosphorylated in, but there is no assertion for organic acids via is modified in. According to this model, an organic acid cannot be modified; however, it can be phosphorylated (and thus modified) in an organic-acid-phosphorylation event. Our analogous solution is to integrate organic acid phosphorylation into the set of allowed classes of is modified in at the level of organic acids (which first requires adding the slot to organic acids and then adding organic acid phosphorylation as an allowed class); the result of this can be seen in Figure 3c. Now, an organic acid can be modified (or, more specifically, phosphorylated) in an organic-acid-phosphorylation event, and there is no inconsistency.
Since some of the slots were arranged hierarchically, it was necessary to check that the set of allowed classes for the superslots of the slot of the assertion at the level of the subject class of the assertion includes the object class of the assertion, examples of which are illustrated in Figure 3. If the set of allowed classes of a superslot was expanded so as to include the object term of the assertion, as was done for the example in Figures 3b, it was furthermore necessary to recursively check to expand the set of allowed classes of the superslot at the level of the subject class’s superclasses. That is, if the object class was propagated up the slot hierarchy, it had to also be checked to be propagated along the class hierarchy using the superslots, which may result in assertions being made with superslots at the level of superclasses; this is again done to avoid inconsistencies of the type illustrated above. For example, since is modified in was added to organic acids and organic acid phosphorylation was made an allowed class of this slot at this level, it must also be checked that is modified in is also attached to each superclass of organics acids that is within the domain of is modified in and that organic acid phosphorylation is an allowed class of this slot at the level of each of these superclasses. Propagation (finally!) terminated when no expansion of sets of allowed classes was necessary, either along the class or slot hierarchies, or when the tops of the slot hierarchy and the tops of the domain classes of these slots were reached. At this point, all original assertions and their inverse assertions were fully integrated into a Protege frame-based ontology that is consistent in that the set of allowed classes of each slot at each class level is a subset of those at higher (i.e., more general) class levels and at higher slot levels. Furthermore, the ontology is also nonredundant in that each set of allowed classes contains no class that is subsumed by or equal to another class in the set.
2.6 Validation of the Integration of the Assertions into the Ontology
Several scripts were written to test the representation and integration of the assertions. One of these scripts tested that the assertions, originally stored in text files, were properly represented using instances of slot with direct allowed classes, attached to classes via the slot has slots with direct allowed classes. Another script verified that each assertion was integrated along the appropriate class and slot hierarchies. More specifically, it was checked that the slot of each assertion is attached to the subject class and that the object class of the assertion is either an explicit allowed class of the slot or a subclass of at least one of its allowed classes (as it may have been removed due to redundancy). This was also checked at the level of each combination of superclass of the subject class within the domain of the slot in question and each superslot of the slot. Furthermore, it was tested that each explicitly represented allowed class of each slot of each class is justified by the presence of an assertion made using the class or one of its subclasses, the slot or one of its subslots, and the allowed class.
3. Results
Figure 4 shows a screenshot of the top-level concepts of the integration of the GO subontologies, ChEBI, and CTO in Protege; note that at this stage this is only the base ontology resulting from the merging of these ontologies. There are eight direct subclasses of :THING corresponding to OBO terms: biological_process, cellular_component, molecular_function (the roots of the GO subontologies), part of biological_process, part of cellular_component, part of molecular_function (to capture their orphan terms), chemical_ontology (the root of ChEBI), and entity developed from cell (to capture the orphan terms of CTO). The cellular-component subontology of GO has a cell class, under which the direct subclasses of CTO’s root cell (and thus all of their subclasses as well) are placed so that all entities explicitly asserted to be cells are in one location.
Figure 4.

A screenshot of our ontology loaded in Protege showing the top-level classes of our ontology and the metaclasses denoting the types of OBO terms. The terms part of entity developed from cell, part of biological_process, part of cellular_component, and part of molecular_function were created to capture orphan terms. The GO term primary cell septum hydrolysis is selected, and all of its information can be seen in the right pane, including its definition, definition reference, unique identifier, synonym, and its relationship with the term of which it is part (modeled as a slot and allowed class).
The 31 GO-term patterns that were selected from the mined list of patterns can be examined in Table 1. Also included in this table are the 25 slots to which these patterns have been mapped. Note that these numbers are different since multiple patterns can be mapped to a given slot. (Multiple slots could conceivably also be mapped to one pattern, but this has not been done at this stage of the project.)
A total of 6,695 GO terms matched the 31 patterns; the number of terms matching each pattern can be examined in the second column of Table 2. An assertion was predicted for 4,184 of these terms through the mapping of text matching an embedded subpattern of the corresponding pattern to an OBO term, which corresponds to 62% of the matching terms having a predicted assertion. The number of terms matching a given pattern for which an assertion was predicted can be seen in the third column of Table 2. Of the 4,184 predicted assertions, 4,068 (97%) were judged to be correct. (These we consider true positives.) The fourth column of Table 2 lists the number of assertions assessed as correct for each pattern. Assertions involving the matching terms using the 25 relationships that were not predicted but assessed as valid were manually added for an additional 429 terms, which amounts to 6% of all terms matching the patterns. (We consider these false negatives.) The number of these assertions added for each pattern (as well as this number as a percentage of the number of terms matching this pattern) can be examined in the fifth column of Table 2. The true positives and false negatives, both of which are integrated into the ontology, sum up to 4,497 assertions (which corresponds to 67% of the total matching terms having an assertion), breakdowns of which can be found in the sixth column of Table 2. With the addition of the inverse assertions, a total of 8,994 assertions using 50 relationships were created ultimately from the presented methodology. Finally, checking to propagate the assertions’ object classes up the class and slot hierarchies resulted in the creation of a total of 20,764 assertions in the form of triples of classes, their slots, and their allowed classes. Figure 5 is a Protege screenshot showing an example term, the ChEBI term carbohydrates, with its enriching links to other OBO terms in the final, integrated ontology.
Table 2.
Patterns and their numbers. In the first column are the patterns, which are written using a simplified syntax, as in Table 1. The number of terms matching each of these patterns is shown in the second column, and the number of these having a predicted assertion, along with this number as a percentage of the number of matching terms (i.e., the second column), is displayed in the third column. The fourth column displays the number of assertions that were judged to be correct for each pattern, along with this number as a percentage of the number of predicted assertions (i.e., the third column). In the fifth column are the number of assertions not predicted but explicitly added for each pattern and this number as a percentage of the number of matching terms for the pattern (i.e., the second column). Finally, the sixth column lists the total number of assertions added (the sum of the fourth and fifth columns) for each pattern along with this number as a percentage of the number of matching terms for the pattern (i.e., the second column). The final row totals each of the last five columns.
| Pattern | Terms | TP+FP | TP | FN | TP+FN |
|---|---|---|---|---|---|
| detection of x | 46 | 1 (2%) | 0 (0%) | 11 (24%) | 11 (24%) |
| negative regulation of x | 526 | 476 (90%) | 476 (100%) | 23 (4%) | 499 (95%) |
| positive regulation of x | 521 | 466 (89%) | 463 (99%) | 22 (4%) | 485 (93%) |
| regulation of x | 566 | 497 (88%) | 497 (100%) | 24 (4%) | 521 (92%) |
| x assembly* | 108 | 50 (46%) | 39 (78%) | 25 (23%) | 64 (59%) |
| x binding* | 693 | 173 (25%) | 167 (96%) | 13 (2%) | 180 (26%) |
| x biosynthesis* | 893 | 577 (65%) | 571 (99%) | 23 (2%) | 594 (66%) |
| x catabolism* | 775 | 596 (77%) | 593 (99%) | 39 (5%) | 632 (82%) |
| x dephosphorylation* | 7 | 4 (57%) | 4 (100%) | 1 (14%) | 5 (71%) |
| x development* | 192 | 13 (7%) | 12 (92%) | 0 (0%) | 12 (6%) |
| x differentiation* | 127 | 66 (52%) | 61 (92%) | 16 (12%) | 77 (61%) |
| x disassembly* | 22 | 11 (50%) | 11 (100%) | 3 (14%) | 14 (64%) |
| x division* | 25 | 9 (36%) | 9 (100%) | 8 (32%) | 17 (68%) |
| x docking* | 10 | 2 (20%) | 2 (100%) | 4 (40%) | 6 (60%) |
| x fermentation* | 30 | 20 (67%) | 11 (55%) | 9 (30%) | 20 (67%) |
| x fission* | 3 | 1 (33%) | 1 (100%) | 2 (67%) | 3 (100%) |
| x formation* | 232 | 46 (20%) | 42 (91%) | 4 (2%) | 46 (20%) |
| x fusion* | 44 | 9 (20%) | 5 (56%) | 21 (48%) | 26 (59%) |
| x inheritance* | 8 | 7 (88%) | 7 (100%) | 1 (12%) | 8 (100%) |
| x lumen* | 17 | 8 (47%) | 7 (41%) | 9 (53%) | 16 (94%) |
| x maturation* | 20 | 6 (30%) | 6 (100%) | 2 (10%) | 8 (40%) |
| x membrane* | 177 | 54 (30%) | 49 (91%) | 29 (16%) | 78 (44%) |
| x metabolism* | 896 | 671 (75%) | 664 (99%) | 53 (6%) | 717 (80%) |
| x modification* | 68 | 30 (44%) | 12 (40%) | 5 (7%) | 17 (25%) |
| x oxidation* | 42 | 31 (74%) | 25 (81%) | 5 (12%) | 30 (71%) |
| x phosphorylation* | 49 | 27 (55%) | 12 (44%) | 2 (4%) | 14 (28%) |
| x proliferation* | 21 | 9 (43%) | 9 (100%) | 4 (19%) | 13 (62%) |
| x reduction* | 23 | 4 (17%) | 3 (75%) | 1 (0.4%) | 4 (17%) |
| x salvage* | 33 | 28 (85%) | 26 (93%) | 0 (0%) | 26 (79%) |
| x secretion* | 40 | 18 (45%) | 18 (100%) | 0 (0%) | 18 (45%) |
| x transport* | 481 | 274 (57%) | 266 (97%) | 70 (14%) | 336 (70%) |
| total | 6695 | 4184 (62%) | 4068 (97%) | 429 (6%) | 4497 (67%) |
Figure 5.

A Protege screenshot showing the enriching links using new, specific relationships for the ChEBI term carbohydrates. For example, this term has been linked to the GO biological-process term carbohydrate transport via the relationship is transported in, stating that a carbohydrate can be transported in a carbohydrate-transport event. Note that there are many allowed classes of these slots that were not directly asserted at the level of carbohydrates but were propagated up to this level to maintain overall semantic consistency of the integrated ontologies, as described in Section 2.5. For example, NADP binding is an allowed class of the slot binds in at this level, stating that a carbohydrate can bind to another entity in an NADP-binding event. The upward propagation of this constraint was needed because NADP(+), the level at which this constraint was directly asserted, is a subclass of carbohydrates in ChEBI, but NADP binding is not a subclass of carbohydrate binding in GO. Without this propagation, there would be a semantic inconsistency in that a carbohydrate could not bind to another entity in an NADP-binding event, yet a molecule of NADP, which is a carbohydrate, could bind to another entity in an NADP-binding event.
4. Discussion
Because Protege lacks system constructs to represent expressions involving unions and intersections of classes, allowed classes of slots that have been inherited from ancestor terms cannot sometimes be represented adequately. Consider a relatively simple case involving two classes a and b with a common subclass c (see Figure 6). a has been directly linked to class x via slot s, and b has been directly linked to class y via s. Since c has not been directly linked to any class via s, it should inherit this slot and its allowed classes from its ancestors. In this case, the allowed classes of s at the level of c would ideally be set to x y. That is, if the value of s for an instance of a must be an instance of x and if the value of s for an instance of b must be an instance of y, then the value of s for instance of c (which is also an instance of a and of b) must be an instance of x and of y. In Protege, when a class inherits multiple allowed classes for the same slot from multiple superclasses (as in the example just presented), Protege arbitrarily chooses the set of allowed classes from only one of the superclasses (both in the GUI and the API). One example of this occurs in the frame of the biological-process term androgen catabolism. One parent term of this term, steroid catabolism, was asserted to be linked to the ChEBI term steroids via the slot results in breakdown of; thus a steroid-catabolism event can result in the breakdown of a steroid. Another parent, hormone catabolism, was asserted to be linked to the ChEBI term hormones; analogously, a hormone-catabolism event can result in the breakdown of a hormone. androgen catabolism was not asserted to have a value for this slot, but since it is a subclass of steroid catabolism and of hormone catabolism, it should inherit the allowed classes of its parents for this slot. Ideally, the allowed class of results in breakdown of at the level of androgen catabolism would be the intersection of steroids and hormones; this would state that an androgen-catabolism event can result in the breakdown of something that is a steroid and a hormone. However, Protege arbitrarily chooses steroids as the allowed class of the slot at this level. This is a difficult problem since it requires arbitrarily complex expressions of unions and intersections of classes, as the patterns of inheritance can be much more complex than that seen in this example. One advantage of using a description-logic-based language such as OWL [14] is that its constructs could be used to exactly create such expressions, or an associated reasoner could be used to enforce such constraints when populating a model.
Figure 6.
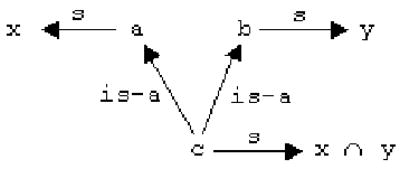
A graph exemplifying inheritance of multiple sets of allowed classes. If any value of slot s for an instance of class a must be an instance of x, and if any value of s for an instance of class b must be an instance of y, then any value of s for an instance of c, which is also an instance of a and of b, should be an instance of x and of y.
Although Protege does not have system constructs to use to build such logical expressions, it is possible to explicitly represent such expressions using frames. Classes representing these expressions could be created and placed appropriately within the ontology (e.g., the class representing the union of classes x and y would directly subsume each of these classes), but this would require the construction of a reasoner to perform this automatic classification whenever a change is made to the ontology, a task that is beyond the scope of this project and would be better handled by existing reasoners of logic-based representations should we move to such a representation. Inherited allowed classes therefore are not represented at all, apart from that which Protege infers by default, including its arbitrary choices of allowed classes when inheriting from multiple parents. As a result of this, the inherited allowed classes of a given slot at the level of a given class (if there are any) must be computed dynamically and returned to the user upon querying. To determine if there are any allowed classes of a given slot at a given class level, it must be determined if there are any allowed classes of the slot at higher class levels that are not represented at the given class level and that do not subsume any of those that are represented at the given class level (since these are overriding, narrowing restrictions on the slot at a lower class level). It must also be checked that an allowed class of the slot at a higher class level indeed comes from a higher class level (i.e., from an assertion made at a higher class level) and not from a lower class level from another subgraph of the ontology through the previously described method of upward propagation, as an allowed class should not be obtained from a class in a separate subgraph.
Although the methodology presented in this paper relies on a frame-based representation, aspects of these methods are likely relevant for other representations of enriched OBO ontologies. In particular, checking to propagate object classes up the hierarchies of the slot and subject class may be necessary to ensure the consistency of the ontology, depending on the knowledge model of the representation used. For example, if an OWL-based approach is used in Figure 2b, employing an OWL allValuesFrom constraint (which is most semantically similar to a nonrequired slot in Protege) to represent the relationship between OWL classes, the propagation should still be performed. Using this OWL-based representation, the semantics of the underlying assertions of the example in Figure 2b are essentially the same: Nitrogen compound metabolism can only result in the metabolism of a nitrogen compound, while alkaloid metabolism, which is a kind of nitrogen compound metabolism, can only result in the metabolism of an alkaloid, which is not a kind of nitrogen compound. A reasoner may not detect this intuitive inconsistency, as it is possible to satisfy this model by, when creating an instance of alkaloid metabolism, setting the value of results in metabolism of to an individual that is a member of both nitrogen compounds and alkaloids. However, this is sidestepping the issue that there is a fundamental discrepancy between the GO and ChEBI ontologies in that alkaloid metabolism is subsumed by nitrogen compound metabolism in GO, which implies that alkaloids are nitrogen compounds, while alkaloids is not subsumed by nitrogen compounds in ChEBI. A corresponding argument can be made using the existential OWL someValuesFrom constraint. It is better to explicitly structure the ontology taking such discrepancies (which may be intended or unintended) into account rather than putting the onus on the user to figure out how to correctly use an ontology so as to ensure consistency. Furthermore, although OBO ontologies were used in this project, the methodology for the integration of assertions using existing terms presented here is applicable to any frame-based ontology.
5. Related Work
In their lexical analysis of the compositionality of GO terms, Ogren et al. searched for GO terms that contain other GO terms as proper substrings and claimed that these occurrences often imply derivational relationships between the terms. They noted substrings that are not GO terms themselves but occur frequently, calling them complements (which correspond to the invariant parts of our patterns); the subset of these they believed semantically meaningful were termed derivational phrases (which correspond to the invariant parts of those patterns that were considered reasonably semantically consistent, including chosen for this project). In addition to finding patterns with derivational phrases, our methodology results in the creation of specific assertions using these patterns and the integration of these assertions into a fully consistent ontology.
There have been several other efforts to attempt to programmatically discover associations between OBO terms. Bada et al. mined the Gene Ontology Annotation database for over 600,000 associations among GO terms in the sense that the two terms of a given association had been used together in at least one GO annotation [15]. Unlike in this work, their associations were used not to suggest formal links between terms in ontologies but to aid in the manual annotation of genes and gene products by suggesting terms to an annotator based on term that had already been applied to the gene or gene product being annotated. Other efforts have produced associations that have been claimed to be bases for linking currently unlinked terms of OBO ontologies: Bodenreider et al. surveyed three nonlexical approaches to identify associations between GO terms and compared these approaches with each other and with lexical approaches, finding 7,665 associations from at least one of the nonlexical approaches, a small fraction of which were suggested by more than one approach and even a smaller fraction of which were also suggested by lexical approaches [16]; this was primarily an effort to evaluate the coverage of these methodologies, and the generated associations did not specify the relationships that linked the associated terms and were not checked for semantic validity by a domain expert, as has been done in our project. Burgun and Bodenreider looked for embedded ChEBI terms within GO terms to suggest 771,302 associations between GO terms; however, there also was neither specification of relationship among associated terms nor checking of semantic validity, and the large majority of these proposed associations are likely indirect in that an associated pair of terms would not be directly linked in an ontology but instead connected by a path of relationships. (For example, one of the resulting associations in this study is that between the GO terms uronic acid metabolism and uronic acid transporter activity as a result of their mutual inclusion of the ChEBI term uronic acid, but it is unlikely that an ontologist would directly link these two GO terms to each other.) Kumar et al. mined the TIGR database, applying association-rule induction to produce associations between pairs of terms from different GO subontologies with a calculated support (the fraction of all TIGR annotation sets for which the relevant association rule is correct) and confidence (the ratio of the number of annotation sets that contain all terms in the rule to the number of annotation sets that contain all terms in the antecedent of the rule) for each association [17]; however, there is no semantic assessment apart from these statistics and also no specification of relationships among associated terms, and it is not certain that the fact that a given pair of terms was used together in annotation sets to an extent beyond chosen support and confidence thresholds necessarily implies that the terms should be directly linked in an ontology (although the results of this method are likely to be a valuable source of suggested associations). Johnson et al. evaluated several lexical techniques to find relationships between GO terms and terms from ChEBI, CTO, and the BRENDA tissue ontology, resulting in 91,385 binary associations; they acknowledge that they have not specified relationships for these associations and also that many of the associations are likely indirect [18]. Since relationships are used to directly link entities in ontologies, direct associations between terms (such as the assertions presented in this paper, all of which are direct) are more useful to ontologists than indirect ones, which merely indicate that a given pair of entities are in some way connected by at least one path of links in the conceptual network. A variety of lexical and nonlexical methods such as those presented above may be useful for suggesting associations between terms for the purpose of linking terms in ontologies, but these efforts should be focused on specific relationships deemed appropriate and useful by modelers to avoid a preponderance of vague, indirect relationships that may result otherwise. Furthermore, any programmatically suggested association between terms must be evaluated by domain experts before its addition to an ontology.
The two efforts most comparable to that presented here are the GONG and Obol projects. In the former, Wroe et al. converted GO and a subset of the MeSH chemical ontology into a description-logic format (specifically, DAML+OIL) and analogously created 250 formal definitions of GO metabolism, biosynthesis, and catabolism terms through the addition of restrictions involving corresponding MeSH-chemical concepts. Furthermore, they present a methodology for an evolution toward a more formal GO using such a description-logic-based approach. This study is more restricted than ours in that a relatively small number of associations using only a few types of relationships were added and in that they were manually added. However, they were able to take advantage of a description-logic reasoner to help to classify terms, which resulted in the inference of 22 new is-a links between terms in this subgraph of metabolism concepts, 17 of which were ultimately added into GO. These hierarchical changes inferred by the reasoner likely result from the discrepancies between ontological subgraphs that have been noted in our study. Several examples of the description logics’s capability of being used to create formal term definitions involving more complex expressions (e.g., quantified existential restrictions) were presented with the caveat that the effort in creating such definitions was time- and labor-intensive.
In a subsequent OWL-based GONG study, Aranguren focused on the binding, transport activity, and metabolism subontologies of GO. In a methodology similar to that of ours for creating assertions, regular expressions were used to capture terms in these subontologies, and embedded chemical names were extracted, normalized using a UMLS tool, and matched to MeSH concepts. Existential restrictions using four relationships were then created linking the GO process terms to the matched MeSH chemical terms, after which a reasoner was enacted to classify all of the terms. Instead of reporting the numbers of predicted restrictions and of those assessed to be valid, this study focused on the numbers of GO terms for which a changed placement in the hierarchy was predicted and of those assessed to be valid by the author and by GO curators. For example, for binding terms (which is the only subontology for which a complete list of numbers is provided), 17% of the terms were predicted to have a change in hierarchy placement, 8% were accepted by the author, and 5% were accepted by GO curators. As in their previous study, discrepancies between corresponding concepts in corresponding ontologies are the likely sources of the hierarchical changes inferred by the reasoner. For example, from the fact that Neurotransmitters subsumes Glutamates in MeSH, it was inferred that neurotransmitter binding should subsume glutamate binding in GO. Suggesting a resolution to a discrepancy by a change in the hierarchical arrangement of classes hierarchy is extremely valuable, but if the suggestion is not accepted (e.g., by the ontology curators), the discrepancy continues to exist. Rather than suggesting a way to resolve a discrepancy, we take the discrepancy into account and ensure consistency between the subgraphs by incorporating relevant terms into the sets of allowed classes of slots. If this is later assessed a real discrepancy and resolved through editing of the ontology, our methodology can be used to recompute the sets of allowed classes of slots of the ontology, using its new structure.
Mungall proposed that OBO term names are so highly regular that they can be considered valid sentences of an underlying language he named Obol. A vocabulary of atomic terms was constructed from the tokenization of OBO terms, and the formal language was represented as a set of Prolog production rules containing terminal symbols and nonterminal symbols that decompose to combinations of terminal symbols and/or other nonterminal symbols. Formal definitions of terms were then created from a syntactic parse of each term, where each parse level results in an Aristotelian genus (category) and differentia (a necessary and sufficient condition that distinguishes the term from other terms in the genus), which are given values from the atomic vocabulary. The differentiae correspond to our slots, and the differentiae values correspond to the allowed classes of the slots, although the semantics of these two types of statements are different. A number of the formal definitions seem to be semantically inconsistent with the natural-language definitions. For example, actin cortical patch assembly is given a differentia and value of occurs_in = actin cortical patch, which seems to state that actin cortical patch assembly takes place in an actin cortical patch, yet the former’s natural-language definition is the “assembly of an actin cortical patch, a discrete actin-containing structure found at the plasma membrane of fungal cells”. Many other definitions use differentiae that are given values that correspond to no existing OBO term (e.g., conjugant formation was given a differentia and value of directly_involves_anatomical = conjugant, which is not a valid term). Although we have not made assertions involving concepts we could not find as existing terms, we do believe this could be useful for the suggestion of terms to ontologies, but formal links should not be created until needed terms exist, obviously. Some information seems to be inconsistently represented (e.g., mitochondrial citrate transport uses the differentia in for the location, while plasma membrane pyruvate transport uses occurs_in), while some of the differentiae seem to us suspect or ambiguous (e.g., and, type). We have arranged our relationships into a hierarchy, which was done in the Obol effort; furthermore, we have a methodology to ensure that all created assertions are consistent throughout the ontology. Nevertheless, Obol is an impressive effort, and full parsing of OBO terms as performed in Obol could potentially extend the assertions produced by the simpler patterns described here.
6. Conclusions
The work presented in this paper is an effort to enrich five OBO ontologies through the addition of large numbers of links between existing terms using 50 specific relationships that do not currently exist in the ontologies. A frame-based ontology that incorporates the GO subontologies, ChEBI, and CTO using a representation of relationships (including GO’s part_of and CTO’s develops_from) that we believe to be more semantically valid than that of previous frame-based efforts was created. A list of patterns found in GO terms was formed by manually selecting from a list mined from GO terms, and these patterns were manually mapped to relationships such that a concept that maps to the text matching a given pattern has a significant probability of being related to a concept that maps to the text matching an embedded subpattern via the relationship. We have shown that carefully constructed patterns can be used to create reasonable assertions in that a large majority of the assertions suggested by this methodology were judged to be valid. Nearly 4,500 primary assertions were created, which, together with their nearly 4,500 inverse assertions, were added to the ontology. Linking previously unlinked terms can lead to semantic inconsistencies due to discrepancies between subgraphs of the ontology; our methodology programmatically adds these assertions and propagates them up the class and slot hierarchies such that each assertion is consistent with respect to more general class and slot levels and such that there is no redundancy within a given slot. The ontology was tested in that each assertion was integrated into the set of allowed classes of a slot at all appropriate class and slot levels and, conversely, in that each allowed class of each slot of each class is justified by the existence of an assertion at an appropriate level.
Others have expounded on the advantages that OBO ontologies with more links and more types of links between terms could provide to biomedical researchers. Ours is one approach to this quest, and we have addressed issues that we believe likely to be confronted in other approaches, particularly the need to integrate an arbitrarily large set of assertions into a consistent, unified ontology.
References
- 1.Ashburner M, Mungall CJ, Lewis SE. Ontologies for Biologists: A Community Model for the Annotation of Genomic Data. Proceedings of the Cold Spring Harbor Symposia on Quantitative Biology. 2003:227–236. doi: 10.1101/sqb.2003.68.227. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarkis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogren PV, Cohen KB, Acquaah-Mensah GK, Eberlein J, Hunter L. The Compositional Structure of Gene Ontology Terms. Proceedings of the Pacific Symposium on Biocomputing. 2004:214–225. doi: 10.1142/9789812704856_0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgun A, Bodenreider O. An ontology of chemical entities helps identify dependence relations among Gene Ontology terms. Proceedings of the First International Symposium on Semantic Mining in Biomedicine. 2005 [Google Scholar]
- 5.Wroe CJ, Stevens RD, Goble CA, Ashburner M. A Methodology to Migrate the Gene Ontology to a Description Logic Environment Using DAML+OIL. Proceedings of the Pacific Symposium on Biocomputing. 2003:624–636. doi: 10.1142/9789812776303_0058. [DOI] [PubMed] [Google Scholar]
- 6.Aranguren ME. Improving the structure of the Gene Ontology. Msc dissertation. 2004 [Google Scholar]
- 7.Mungall CJ. Obol: integrating language and meaning in bio-ontologies. Comparative and Functional Genomics. 2004;5:509–520. doi: 10.1002/cfg.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. http://www.ebi.ac.uk/chebi/
- 9.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biology. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noy NF, Fergerson RW, Musen MA. The knowledge model of Protege-2000: Combining interoperability and flexibility; Proceedings of the 2nd International Conference on Knowledge Engineering and Knowledge Management (EKAW) 2000. [Google Scholar]
- 11.Lu Z, Firby J, Baumgartner WA, Jr, Cohen KB, Ogren PV, Hunter L. Ontology-driven analysis of complex relationships in biomedical text: Extracting protein transport information from GeneRIFs. Submitted to Bioinformatics [Google Scholar]
- 12. http://staff.washington.edu/ads99/wiki/obo_convert/
- 13.Yeh I, Karp PD, Noy NF, Altman RB. Knowledge acquisition, consistency checking and concurrency control for Gene Ontology (GO) Bioinformatics. 2003;19(2):241–248. doi: 10.1093/bioinformatics/19.2.241. [DOI] [PubMed] [Google Scholar]
- 14. http://www.w3.org/TR/2004/REC-owl-guide-20040210/
- 15.Bada M, Turi D, McEntire R, Stevens R. Using Reasoning to Guide Annotation with Gene Ontology Terms in GOAT. SIGMOD Record. 2004;33(2):27–32. [Google Scholar]
- 16.Bodenreider O, Aubry M, Burgun A. Non-Lexical Approaches to Identifying Associative Relations in the Gene Ontology. Proceedings of the Pacific Symposium on Biocomputing. 2005:91–102. [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar A, Smith B, Borgelt C. Dependence Relationships between Gene Ontology Terms based on TIGR Gene Product Annotations. Proceedings of CompuTerm. 2004:31–38. [Google Scholar]
- 18.Johnson HL, Cohen KB, Baumgartner WA, Jr, Lu Z, Bada M, Kester T, Kim H, Hunter L. Evaluation of Lexical Methods for Detecting Relationships Between Concepts from Multiple Ontologies. Proceedings of the Pacific Symposium on Biocomputing. 2006:28–39. [PMC free article] [PubMed] [Google Scholar]


