Abstract
Covalent binding of biotin to histones participates in heterochromatin formation, cell cycle progression, and the cellular response to DNA breaks. Biotinylation of histones appears to be a reversible process but the identities of enzymes that remove the biotin mark are largely unknown. Our long-term goal is to identify histone debiotinylases in human cells. Here we developed an avidin-based plate assay to quantify histone debiotinylase activities in nuclear extracts. This assay is an essential first step in purifying and identifying histone debiotinylases from human cells. Using this assay we demonstrated that debiotinylation of histones depends on temperature and pH, consistent with enzyme catalysis. Experiments with purified histones, proteases, and protease inhibitors provide evidence that removal of the biotin mark from histones is mediated by debiotinylases rather than proteases. Activities of histone debiotinylases varied among human tissues: colon = lung > placenta = liver > lymphoid cells. The assay proved useful to monitor activities of putative histone debiotinylases during their partial purification from cells. Collectively, this assay is a useful tool for investigating histone debiotinylases in human tissues.
Keywords: Avidin, biotin, biotinidase, debiotinylation, histones
1. Introduction
The classical role of biotin in intermediary metabolism is to serve as a covalently bound coenzyme for acetyl-CoA carboxylase α, acetyl-CoA carboxylase β, pyruvate carboxylase, propionyl-CoA carboxylase, and 3-methylcrotonyl-CoA carboxylase [1]. These enzymes play essential roles in the metabolism of fatty acids, glucose, and amino acids. In addition, evidence has been provided that the activity of cell signals such as biotinyl-AMP, NF-κB, Sp1, Sp3, and receptor tyrosine kinases depends on biotin [2-5]. Consistent with these observations, biotin and its catabolites affect the expression of greater than 2000 genes in human lymphocytes and liver cells [6-8].
More recently, the quantity and quality of biological functions of biotin has been expanded substantially by the demonstration that histones H1, H2A, H2B, H3, and H4 contain covalently bound biotin [9,10]. The following biotinylation sites have been identified: K9, K13, K125, K127, and K129 in histone H2A [11], K4, K9, and K18 in histone H3 [12], and K8 and K12 in histone H4 [13]. Biotinylation of histones is mediated by holocarboxylase synthase [14] and biotinidase [9]. Evidence has been provided that biotinylation of histones plays a role in cell proliferation [10], the cellular response to DNA damage [15], mitotic condensation of chromatin [16], and heterochromatin structures and gene silencing (Camporeale et al., submitted). Collectively, binding of biotin to histones participates in epigenetic events that are crucial for maintaining genomic stability and chromatin structure [17].
Amino acid residues that are targets for covalent modification in histones are exposed at the nucleosomal surface in chromatin [18,19]. The lysine residues that are targets for biotinylation are no exception to this rule [12,13,20]. Circumstantial evidence has been provided that cells possess the tools needed to remove the biotin mark from these lysine residues; enzymatic debiotinylation of histones is a feasible mechanism to modify chromatin structure in response to changes in the cellular environment. For example, in JAr choriocarcinoma cells, K12 in histone H4 is rapidly debiotinylated in response to double-strand breaks in DNA [15]. Likewise, K12 in histone H4 is debiotinylated at the interleukin-2 locus in response to mitogenic stimulation of lymphoid cells (Camporeale et al., submitted). There is precedence for the notion that removal of histone modifications plays a critical role in the regulation of chromatin structure. For instance, removal of acetyl residues by deacetylases is a known hallmark of gene silencing [21].
The identities of enzymes capable of debiotinylating histones are largely unknown. Evidence has been provided that biotinidase might catalyze both biotinylation [9] and debiotinylation of histones [22]. However, it remains uncertain as to how the same enzyme may act as a biotinyl transferase in one case and as a debiotinylase in another. It has been proposed that regulation of biotinidase is achieved by interactions with chromatin-associated proteins, posttranslational modifications and alternative splicing, and substrate availability [17]. Note that we have detected biotinidase in human cell nuclei [11], whereas other groups suggest that biotinidase is primarily an extranuclear protein [23]. The identification of the exact role of biotinidase in histone debiotinylation awaits further analysis and also depends on the availability of a reliable assay for debiotinylase activity.
A long-term goal of our studies is to identify histone debiotinylases in human cells. As an important first step towards achieving this goal we have developed a microtiter plate assay to quantify activities of histone debiotinylases in nuclear extracts. Here, we provide data to validate this assay and to provide first insights into debiotinylase activities in human tissues. Importantly, we demonstrate that this assay is a useful tool to monitor debiotinylase activities during enzyme purification.
2. Materials and methods
2.1. Cell culture
The following cell lines were obtained from American Type Culture Collection (Manassas, VA): HepG2 hepatocarcinoma cells, JAr choriocarcinoma cells, Jurkat lymphoma cells (clone E6-1), and NCI-H69 small cell lung cancer cells. HCT116 epithelial cells derived from a human colorectal carcinoma were provided by B. Vogelstein (Johns Hopkins University, Baltimore, MD). Cells were cultured in humidified atmosphere (5% CO2 at 37°C) as described [8,24-26]. Cell viability was monitored periodically using the Trypan blue exclusion test [27].
2.2. Protein extracts
Proteins were extracted from cell nuclei and cytoplasm by using the Nuclear Extract Kit (Active Motif, Carlsbad, CA) according to the manufacturer’s instructions. Protein concentrations were determined using the bicinchoninic acid method (Pierce, Rockford, IL).
Quantitation of histone debiotinylase activity
First, histone H1 from calf thymus (Calbiochem, La Jolla, CA) was biotinylated enzymatically [13] to produce substrate for histone debiotinylases in subsequent assays. Briefly, 1 mg of histone H1 was dissolved in a mixture of 0.6 ml of human plasma (as a source of biotinyl transferase), 0.4 ml of 750 μM biocytin (biotinyl-ε-lysine, as a source of biotin), and 19 ml of 50 mM Tris buffer [pH 8.0]; the mixture was incubated at 37°C for 45 min in a waterbath [13]. In previous studies we confirmed covalent binding of biotin to histones by using HPLC and mass spectrometry [13]. Second, 96-well plates were coated with biotinylated histone H1 as follows. Thirty milliliters of 50 mM carbonate buffer [pH 9.6] were added to the histone solution from step 1, and 100-μl aliquots of the mixture were dispensed into 96-well plates for overnight coating at 4°C. Coating efficiency depended substantially on the brand of plate used. We obtained best results by using Falcon Microtest plates (Becton Dickinson, Franklin Lakes, NJ) but readers are encouraged to test various brands. Next, wells were washed with phosphate-buffered saline (PBS)1 [pH 7.4] and blocked with 200 μl of 0.1% bovine serum albumin (w/v) and 0.05% Tween-20 (v/v) in PBS [pH 7.4] at 4°C for at least 4 h. Activities of histone debiotinylases were quantified as follows. Coated and blocked plates were washed twice using 200 μl of PBS. Nuclear or cytoplasmic extracts were diluted with water to produce a concentration of 2.5 μg of protein/50 μl. Fifty microliters of diluted extracts were mixed with 100 μl of 50 mM Tris buffer [pH 7.4], and samples were transferred to microwell plates to initiate the enzymatic debiotinylation of histones; incubation times, temperatures, and variations of buffers were as provided in Results. Typically, protein-free Tris buffer was used as a negative control but other controls were also tested (see below). Debiotinylation of histones was terminated by washing the plates twice with 200 μl of PBS. Histone-bound biotin remaining in the plates was probed using 100 μl of avidin-conjugated horseradish peroxidase [10 μg/l in PBS containing 0.1% BSA (w/v)] at room temperature for 1 h. Plates were washed twice using 0.05% Tween-20 in PBS (v/v). Bound horseradish peroxidase was quantified using 100 μl of SureBlue TMB [3,3′,5,5′-tetra-methylbenzidine] Microwell Peroxidase Substrate (KPL, Inc.; Gaithersburg, MD) at room temperature for 30 min; the reaction was terminated by adding 100 μl of TMB Stop Solution (KPL, Inc.; Gaithersburg, MD) or 1 N hydrochloric acid. The absorbance was read at 450 nm in an Emax Microplate reader (Molecular Devices, Sunnyvale, CA). Calibration curves were generated by incubating avidin-conjugated horseradish peroxidase (up to 1.4 fmol/well) in histone-free plates with 100 μl of TMB substrate. Calibration was based on the assumption that on average one molecule of avidin is conjugated to two molecules of horseradish peroxidase, producing a molecular weight of 147 kDa. One unit of histone debiotinylase activity is defined as 1 pmol of biotin released mg protein-1 15 min-1.
2.3. Proteolytic digestion of histone H1
Theoretically, proteases present in tissue extracts could non-specifically cleave histone H1 in the wells and introduce artifacts in the histone debiotinylases assay. To evaluate potential proteolytic activities, 1 mg of histone H1 was dissolved in 100 μl of 20 mM sodium acetate buffer [pH 4.5]. A volume of acetate buffer containing 4 μg of histone H1 was mixed with 10 μl of 50 mM Tris buffer [pH 7.4] and 7 μl of nuclear extract (containing 20 μg of protein); samples were incubated at 37°C for 20 min. The following controls were tested: (i) histone H1 incubated without nuclear extract, and (ii) histone H1 incubated with 6.25 ng trypsin with or without 20 mM of the trypsin inhibitor phenylmethylsulfonylfluoride (PMSF). Reactions were terminated by heating the samples with an equal volume of gel loading buffer (Invitrogen, Carlsbad, CA) at 72°C for 10 min. Proteins were resolved using 4 -12% Bis-Tris gels (Invitrogen) as described [10] and were visualized using Coomassie blue.
2.4. Partial purification of debiotinylases from Jurkat cell nuclei
Whole cell protein extracts were prepared from Jurkat cells by using the hypotonic buffer from the above Nuclear Extract Kit (Active Motif, Carlsbad, CA) according to the manufacturer’s instructions; extracts were prepared in the presence of protease inhibitor cocktail. Proteins were partially enriched by fractional ammonium sulfate precipitation [28]; ammonium sulfate was removed by two changes against 1000 volumes of 50 mM Tris buffer, pH 7.4. Peak debiotinylase activities were detected in the 30-75% ammonium sulfate fraction, and this fraction was used for further protein purification. Proteins were resolved by anion-exchange chromatography (Q Sepharose; Amersham Pharmacia Biotech, Uppsala, Sweden) and eluted by using 0.5 M NaCl in 50 mM Tris, pH 7.4.
2.5. Statistics
Homogeneity of variances among groups was confirmed using Bartlett’s test [29]. Significance of differences among groups was tested by one-way ANOVA. Fisher’s Protected Least Significant Difference procedure was used for posthoc testing [29]. Student’s paired t-test was used for pairwise comparisons. StatView 5.0.1 (SAS Institute; Cary, NC) was used to perform all calculations. Differences were considered significant if P< 0.05. Data are expressed as mean ± SD.
3. Results
3.1. Calibration and linearity of the histone debiotinylase assay
The apparent oxidation of TMB increased linearly up to 0.7 fmoles of avidin-horseradish peroxidase per well, as judged by the absorbance at 450 nm (Fig. 1). Subsequent histone debiotinylation assays were calibrated using avidin standards from within the linear range. For purposes of calibration it was assumed that one molecule of avidin binds four molecules of biotin [30]. This might slightly overestimate the amount of biotin released by histone debiotinylases, given that not all biotin-binding sites in avidin might participate in biotin binding due to spatial effects [31].
Fig. 1.
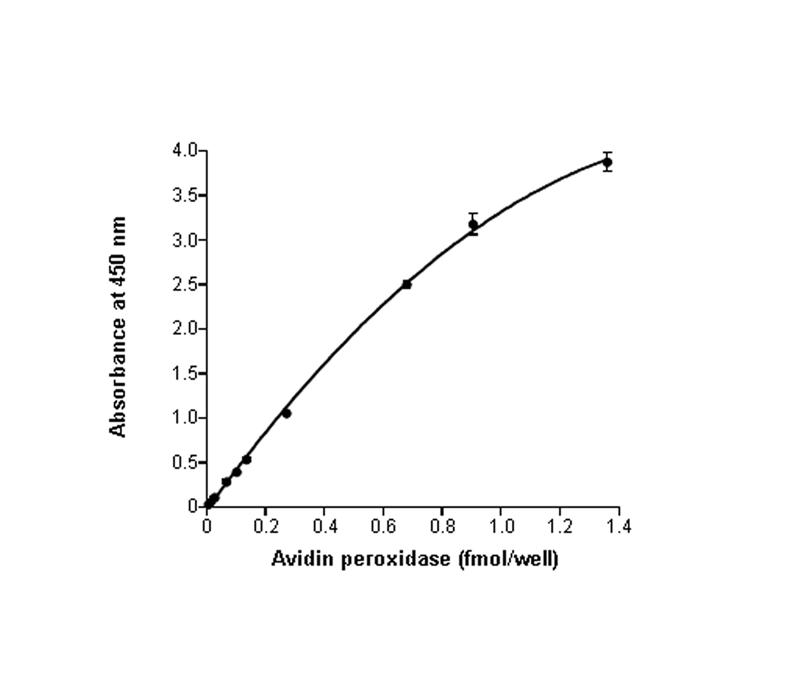
Spectrophotometric quantitation of TMB oxidation. The oxidation of TMB in SureBlue TMB Microwell Peroxidase Substrate (100 μl/well) increased linearly up to 0.7 fmol/well avidin-horseradish peroxidase.
Incubation of 96-well plates with nuclear extracts from NCI-H69 cells caused a time- and protein-dependent release of biotin from histone H1. This is consistent with the presence of histone debiotinylases in human cell nuclei. The release of biotin was linear up to about 5 μg of nuclear protein per well (Fig. 2). If the mass of protein exceeded 5 μg per well, we observed an artificial rapid “debiotinylation” of histones during the first 5 minutes of incubation. This apparent rapid debiotinylation could not be blocked by heat inactivation of nuclear extracts or by incubation of plates at 4°C and, hence, was not an enzyme-mediated process. In contrast, if plates were incubated with up to 5 μg protein/well, histone debiotinylation exhibited all characteristics of enzyme-mediated processes (see below). Consequently, all assays described below were conducted using 2.5 μg of nuclear protein per well for 15 min. In a typical experiment the activity of histone debiotinylases in NCI-H69 nuclei equaled 0.7 ± 0.2 units. The signal-to-noise ratio of our assay was approximately 24 to 1.
Fig. 2.
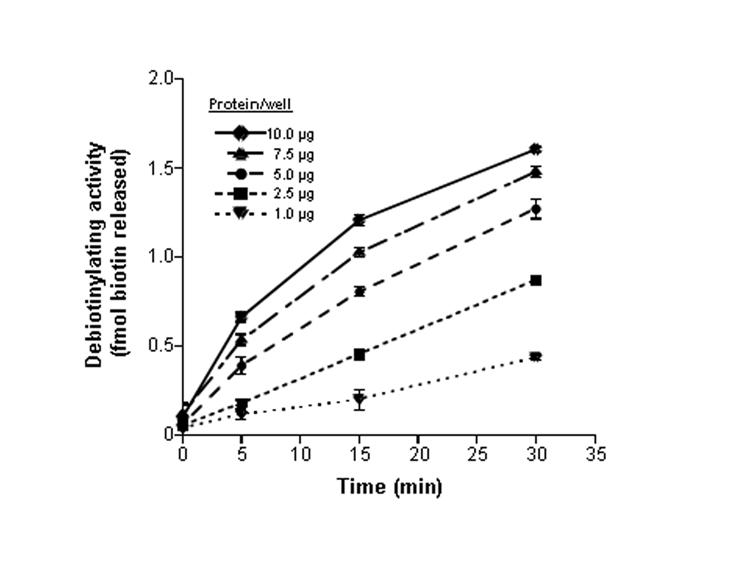
Histone debiotinylation depends on the amount of nuclear protein and incubation time. Histone H1-coated plates were incubated with nuclear protein from NCI-H69 cells and rates of histone debiotinylation were quantified at timed intervals at pH 7.4 and 37°C (n = 3 for each data point).
3.2. Debiotinylation characteristics
Debiotinylation of histones by nuclear extract is an enzyme-mediated process as opposed to being caused by artificial desorption of histone H1 from plastic surfaces, based on the following line of observations. First, debiotinylation of histone H1 by nuclear extracts from NCI-H69 cells was temperature dependent: 0.7 ± 0.03 units at 37°C, versus 0.4 ± 0.02 units at 4°C (P< 0.05; n = 3). Second, heating nuclear extracts at 90°C abolished histone debiotinylase activity. Third, debiotinylation of histone H1 was not detectable if plates were incubated with protein-free nuclear extraction buffer. Fourth, the rate of histone debiotinylation depended on the pH of the incubation buffer (Fig. 3). Here, pH values in the incubation buffer were varied between 3.5 and 10, using buffers based on 100 mM (final concentration) sodium acetate (pH 3.5 -5), 100 mM 2-(N-Morpholino)ethanesulfonic acid (pH 6 -7), and 100 mM Tris (pH 8 -10) buffer. Two pH optima were observed for histone debiotinylation in nuclei, consistent with multiple enzymes mediating release of biotin from histones. The first pH optimum was between 4.0 - 4.5. The second pH optimum was between 7.5 - 8.0. The latter is similar to the pH in the nuclear compartment and similar to the pH optimum of the aminohydrolase biotinidase [28]. Given the known role of biotinidase in debiotinylation reactions, the majority of subsequent studies was conducted at pH 7.4.
Fig. 3.
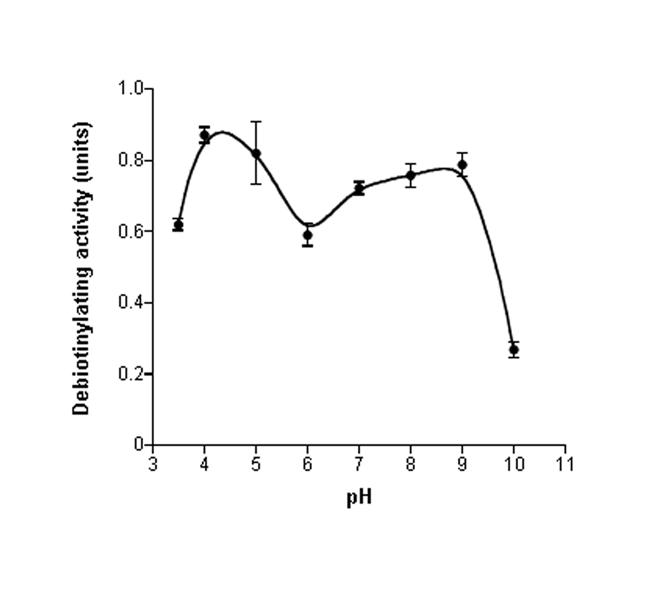
Debiotinylation of histone H1 by nuclear extracts from NCI-H69 cells depended on the pH of the incubation buffer. Rates of debiotinylation were quantified at 37°C for 15 min (n = 3).
3.3. Proteolysis
To exclude potential proteolytic degradation of histone as source of error in our assays, commercial histone H1 was incubated with nuclear extract from NCI-H69 cells, and the electrophoretic pattern of histone H1 was observed in Coomassie blue stained gels. The majority of histone H1 remained intact during incubation with nuclear extract (Fig. 4, lanes 1 and 2). The sample containing both histone H1 and nuclear extract (lane 1) produced multiple extra bands compared with the sample containing only histone H1 (lane 2). These bands represented nuclear proteins rather than breakdown of histone H1. This notion is based on the observation that a sample containing nuclear extract but no supplemental histone H1 produced a staining pattern similar to the sample supplemental with histone H1 (compare lanes 1 and 3). Histone H1 was degraded if incubated with 6.25 ng trypsin (lane 4; positive control); degradation by trypsin was inhibited by using the trypsin inhibitor PMSF (lane 5). Note that the extraction buffer used for preparation of nuclear extracts contains protease inhibitors, consistent with minimal if any proteolysis in the debiotinylation assays. Collectively, we cannot formally exclude the possibility that proteases account for some removal of biotin marks from histones. However, our data strongly suggest that debiotinylases rather that proteases account for histone debiotinylation (see Discussion).
Fig. 4.
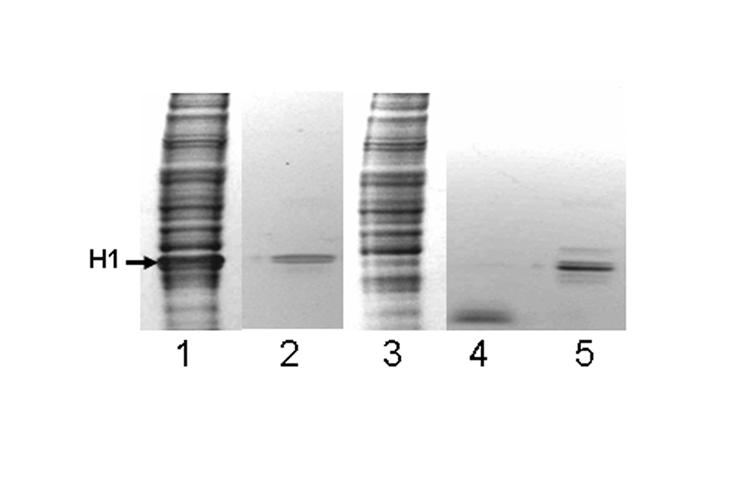
Release of biotin by proteolytic degradation of histone H1 was quantitatively minor. Lane 1 = histone H1 incubated with nuclear extract from NCI-H69 cells (37°C, 20 min); lane 2 = native histone H1 incubated in the absence of nuclear extract; lane 3 = nuclear extract from NCI-H69 in the absence of exogenous histone H1; lane 4 = histone H1 incubated with 6.25 ng trypsin (37°C, 20 min); and lane 5 = histone H1 incubated with trypsin in the presence of 20 mM PMSF (37°C, 20 min). Proteins were resolved by gel electrophoresis and stained with Coomassie blue.
3.4. Tissue distribution and cellular localization
Histone debiotinylases exhibited a tissue-specific pattern of distribution. Of all the tissues tested, NCI-H69 lung cancer cells and HCT116 human colorectal carcinoma epithelial cells exhibited the highest debiotinylase activities (Fig. 5). Debiotinylase activities in HepG2 hepatocarcinoma cells and JAr choriocarcinoma cells were about 50% smaller than in NCI-H69 cells, and activities in Jurkat lymphoid cells were about 70% smaller than in NCI-H69 cells. The activity of histone debiotinylases was greater in cell nuclei compared with cytoplasm. For example, debiotinylase activity was 0.8 ± 0.04 units in nuclei from NCI-H69 cells, but only 0.3 ± 0.03 units in cytoplasmic extracts (P< 0.05; n = 3).
Fig. 5.
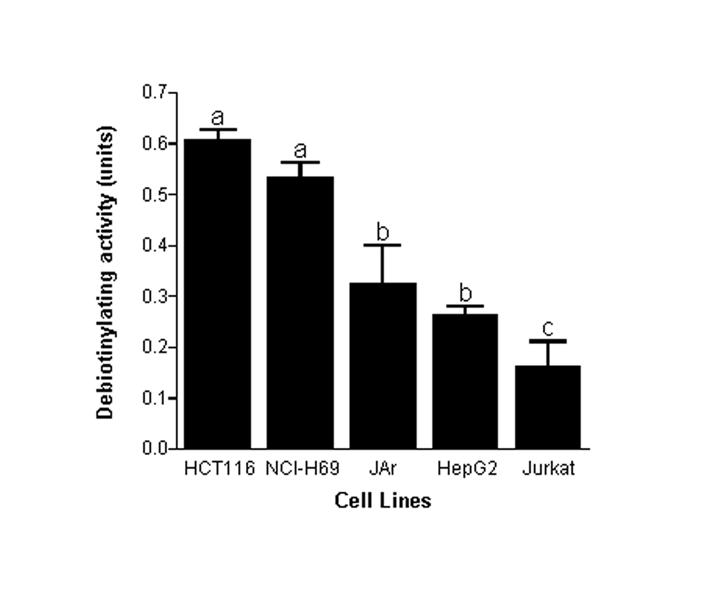
Activities of histone debiotinylases in nuclear extracts from human cells depended on the tissue from which cells originated. Nuclear extracts were prepared from HCT 116 colorectal carcinoma, NCI-H69 small lung cancer, JAr choriocarcinoma, HepG2 hepatocarcinoma and Jurkat lymphoma cells and debiotinylase activities were quantified by plate assay at pH 7.4. a,b,cColumns not sharing the same letter are significantly different (P< 0.05; n = 3).
3.5. Partial purification of histone debiotinylases
The assay developed here is a useful tool to monitor activities of histone debiotinylases during purification of cellular proteins. Here, we used ammonium sulfate precipitation and anion-exchange chromatography to achieve partial purification. Peak debiotinylase activities were detected in fractions obtained by using 30-75% ammonium sulfate, and anion-exchange fractions eluted with 0.5 M NaCl. Overall, we achieved a four-fold purification of human histone debiotinylases (Fig. 6). Purification of histone debiotinylases to homogeneity was beyond the scope of these studies but will be pursued in future investigations. Note that the precipitation of histone debiotinylase in the 30-75% ammonium sulfate fraction was fairly similar to the known enrichment of biotinidase [28], an enzyme suspected to have histone debiotinylase activity [22].
Fig. 6.
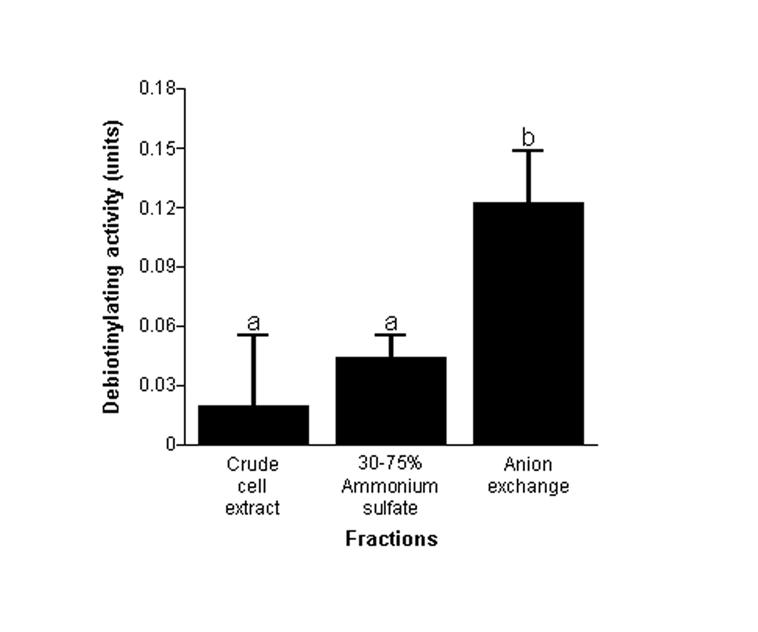
Semi-purification of histone debiotinylases from Jurkat cells. Proteins were fractionated by using 30-75% ammonium sulfate and anion-exchange chromatography (0.5 M NaCl). Specific activities of histone debiotinylase were quantified in crude cell extracts, ammonium sulfate fractions, and fractions collected by anion exchange chromatography. a,bColumns not sharing the same letter are significantly different (P < 0.05; n = 3).
Discussion
Here we present an avidin-based assay to quantify activities of histone debiotinylases. Using this assay we provide evidence (i) that human cell nuclei contain proteins with histone debiotinylase activity; (ii) that debiotinylation of histones is mediated by debiotinylases rather than proteases; (iii) that histone debiotinylases are enriched in the nuclear compartment; and (iv) that the activities of histone debiotinylases are greater in cells derived from lung and colon compared with cells derived from the lymphatic system, liver and placenta. Importantly, we also provide evidence that this assay is a useful tool to achieve our long-term goal, i.e., the identification of histone debiotinylases in human cells.
These findings are biologically relevant, given the following observations from previous studies. First, biotinylation of histones plays a role in cell proliferation [10,14], the cellular response to DNA damage [15], mitotic condensation of chromatin [16], and heterochromatin structures and gene silencing (Camporeale et al., submitted). Abnormal patterns of histone biotinylation might cause some of the events known to be associated with biotin deficiency, e.g., fetal malformations [32]. Histone debiotinylation likely plays an important role in chromatin remodeling events. For example, the biotin mark is rapidly removed from K12 in histone H4 in response to DNA double-stranded breaks [15] and gene activation (Camporeale et al., submitted). Relatively little is known about the enzymes that mediate removal of the biotin mark from histones. The assay presented here breaks ground for future investigations of histone debiotinylases. Second, biotinidase is suspected to play a role in histone debiotinylation [22]. Inborn errors causing biotinidase deficiency are fairly common in humans. The estimated incidence of profound biotinidase deficiency (<10% of normal biotinidase activity) is one in 112,000 live births, and the incidence of partial biotinidase deficiency (<30% of normal biotinidase activity) is one in 129,000 [33]. The combined incidence of profound and partial deficiency is 1 in 60,000 live births; an estimated 1 in 123 individuals is heterozygous for the disorder [33]. Mutations of the biotinidase gene have been well characterized at the molecular level [34-36]. It remains to be determined whether biotinidase deficiency causes abnormalities in chromatin structure. The assay presented here will facilitate these studies.
In the present study, nuclear histone debiotinylases exhibited two pH optima; the first pH optimum was about 4.0 - 4.5 and the second pH optimum was about 7.5 - 8.0. Notwithstanding the putative role of biotinidase in histone debiotinylation, we cannot formally exclude the possibility that enzymes other than biotinidase also mediate debiotinylation of histones. Biotinidase belongs to the nitrilase superfamily of enzymes, which consists of 12 families of amidases, N-acyltransferases, and nitrilases [37]. Some members of the nitrilase superfamily (vanins-1, -2, and -3) share significant sequence similarities with biotinidase [38]. The assay developed here is likely to generate insights into potential roles of enzymes other than biotinidase in histone debiotinylation and chromatin structure.
We shall point out the following uncertainties associated with the studies presented here. First, proteolytic degradation of histone was monitored by staining gels with Coomassie blue. This assay may lack the sensitivity to detect quantitatively minor proteolysis events. In addition, proteolytic removal of only a few amino acids from histone H1 may cause only a minor shift in electrophoretic mobility that is too small for detection. Second, our assay has the same limitation as many other plate assays for histone modifications: enzyme activities are quantified in an isolated system rather than the chromatin context. We cannot formally exclude the possibility that proteins interacting with debiotinylases cause meaningful alterations of debiotinylase activity. Third, the enzymatic biotinylation of histone H1 during substrate synthesis was likely associated with biotinylation of various sites in histone H1. The identity of these sites is unknown. We are currently in the process of developing biotinylation site-specific assays for histone debiotinylases. These assays target known biotinylation sites in histone H2A, H3, and H4 that have been identified in out previous studies [11-13]. These limitations notwithstanding, we anticipate that the histone debiotinylase assay presented here will permit identification of novel nuclear debiotinylases and help generate insights into biotin-dependent chromatin remodeling events.
Acknowledgments
This work was supported by NIH grants DK 60447 and DK 063945, USDA grant 2006-01540, and by NSF EPSCoR grant EPS-0346476. This paper is a contribution of the University of Nebraska Agricultural Research Division, Lincoln, NE 68583 (Journal Series No. 15166).
Footnotes
- PBS
- phosphate-buffered saline
- PMSF
- phenylmethylsulfonylfluoride
- TMB
- 3,3′,5,5′-tetra-methylbenzidine.
References
- 1.Camporeale G, Zempleni J, Bowman BA, Russell RMs. Present Knowledge in Nutrition. International Life Sciences Institute; Washington, D.C.: 2006. Biotin. (in press) [Google Scholar]
- 2.Solorzano-Vargas RS, Pacheco-Alvarez D, Leon-Del-Rio A. Holocarboxylase synthetase is an obligate participant in biotin-mediated regulation of its own expression and of biotin-dependent carboxylases mRNA levels in human cells. Proc Natl Acad Sci USA. 2002;99:5325–5330. doi: 10.1073/pnas.082097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez-Melendez R, Schwab LD, Zempleni J. Jurkat cells respond to biotin deficiency with increased nuclear translocation of NF-κB, mediating cell survival. Int J Vitam Nutr Res. 2004;74:209–216. doi: 10.1024/0300-9831.74.3.209. [DOI] [PubMed] [Google Scholar]
- 4.Griffin JB, Rodriguez-Melendez R, Zempleni J. The nuclear abundance of transcription factors Sp1 and Sp3 depends on biotin in Jurkat cells. J Nutr. 2003;133:3409–3415. doi: 10.1093/jn/133.11.3409. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Melendez R, Griffin JB, Sarath G, Zempleni J. High-throughput immunoblotting identifies biotin-dependent signaling proteins in HepG2 hepatocarcinoma cells. J Nutr. 2005;135:1659–1666. doi: 10.1093/jn/135.7.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Melendez R, Lewis B, McMahon RJ, Zempleni J. Diaminobiotin and desthiobiotin have biotin-like activities in Jurkat cells. J Nutr. 2003;133:1259–1264. doi: 10.1093/jn/133.5.1259. [DOI] [PubMed] [Google Scholar]
- 7.Wiedmann S, Rodriguez-Melendez R, Ortega-Cuellar D, Zempleni J. Clusters of biotin-responsive genes in human peripheral blood mononuclear cells. J Nutr Biochem. 2004;15:433–439. doi: 10.1016/j.jnutbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Melendez R, Griffin JB, Zempleni J. The expression of genes encoding ribosomal subunits and eukaryotic translation initiation factor 5A depends on biotin and bisnorbiotin in HepG2 cells. J Nutr Biochem. 2006;17:23–30. doi: 10.1016/j.jnutbio.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem Mol Med. 1995;56:76–83. doi: 10.1006/bmme.1995.1059. [DOI] [PubMed] [Google Scholar]
- 10.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–5429. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 11.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J Nutr Biochem. 2006;17:225–233. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, Zempleni J. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–4259. doi: 10.1111/j.1742-4658.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur J Biochem. 2004;271:2257–2263. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 14.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum Mol Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- 15.Kothapalli N, Sarath G, Zempleni J. Biotinylation of K12 in histone H4 decreases in response to DNA double strand breaks in human JAr choriocarcinoma cells. J Nutr. 2005;135:2337–2342. doi: 10.1093/jn/135.10.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kothapalli N, Zempleni J. Biotinylation of histones depends on the cell cycle in NCI-H69 small cell lung cancer cells. FASEB J. 2005;19:A55. [abstract] [Google Scholar]
- 17.Zempleni J. Uptake, localization, and noncarboxylase roles of biotin. Annu Rev Nutr. 2005;25:175–196. doi: 10.1146/annurev.nutr.25.121304.131724. [DOI] [PubMed] [Google Scholar]
- 18.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 19.Wolffe A. Chromatin. Academic Press; San Diego, CA: 1998. [Google Scholar]
- 20.Chew YC, Raza AS, Sarath G, Zempleni J. Biotinylation of K8 and K12 co-occurs with acetylation and mono-methylation in human histone H4. FASEB J. 2006;20:A610. [abstract] [Google Scholar]
- 21.Zhang Y, Fatima N, Dufau ML. Coordinated changes in DNA methylation and histone modifications regulate silencing/derepression of luteinizing hormone receptor gene transcription. Mol Cell Biol. 2005;25:7929–7939. doi: 10.1128/MCB.25.18.7929-7939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballard TD, Wolff J, Griffin JB, Stanley JS, Calcar Sv, Zempleni J. Biotinidase catalyzes debiotinylation of histones. Eur J Nutr. 2002;41:78–84. doi: 10.1007/s003940200011. [DOI] [PubMed] [Google Scholar]
- 23.Stanley CM, Hymes J, Wolf B. Identification of alternatively spliced human biotinidase mRNAs and putative localization of endogenous biotinidase. Mol Genet Metab. 2004;81:300–312. doi: 10.1016/j.ymgme.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J Nutr. 2002;132:887–892. doi: 10.1093/jn/132.5.887. [DOI] [PubMed] [Google Scholar]
- 25.Scheerger SB, Zempleni J. Expression of oncogenes depends on biotin in human small cell lung cancer cells NCI-H69. Int J Vitam Nutr Res. 2003;73:461–467. doi: 10.1024/0300-9831.73.6.461. [DOI] [PubMed] [Google Scholar]
- 26.Crisp, SERH, Camporeale G, White BR, Toombs CF, Griffin JB, Said HM, Zempleni J. Biotin supply affects rates of cell proliferation, biotinylation of carboxylases and histones, and expression of the gene encoding the sodium-dependent multivitamin transporter in JAr choriocarcinoma cells. Eur J Nutr. 2004;43:23–31. doi: 10.1007/s00394-004-0435-9. [DOI] [PubMed] [Google Scholar]
- 27.Zempleni J, Mock DM. Uptake and metabolism of biotin by human peripheral blood mononuclear cells. Am J Physiol Cell Physiol. 1998;275:C382–C388. doi: 10.1152/ajpcell.1998.275.2.C382. [DOI] [PubMed] [Google Scholar]
- 28.Chauhan J, Dakshinamurti K. Purification and characterization of human serum biotinidase. J Biol Chem. 1986;261:4268–4275. [PubMed] [Google Scholar]
- 29.SAS Institute . StatView Reference. SAS Publishing; Cary, NC: 1999. [Google Scholar]
- 30.Green NM. Avidin. Adv Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- 31.Green NM. Methods in Enzymology. Academic Press, Inc.; New York: 1990. Avidin and Streptavidin. [DOI] [PubMed] [Google Scholar]
- 32.Mock DM, Mock NI, Stewart CW, LaBorde JB, Hansen DK. Marginal biotin deficiency is teratogenic in ICR mice. J Nutr. 2003;133:2519–2525. doi: 10.1093/jn/133.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf B. Worldwide survey of neonatal screening for biotinidase deficiency. J Inher Metab Dis. 1991;14:923–927. doi: 10.1007/BF01800475. [DOI] [PubMed] [Google Scholar]
- 34.Moslinger D, Muhl A, Suormala T, Baumgartner R, Stockler-Ipsiroglu S. Molecular characterisation and neuropsychological outcome of 21 patients with profound biotinidase deficiency detected by newborn screening and family studies. Eur J Pediatr. 2003;1621(Suppl):S46–49. doi: 10.1007/s00431-003-1351-3. [DOI] [PubMed] [Google Scholar]
- 35.Laszlo A, Schuler EA, Sallay E, Endreffy E, Somogyi C, Varkonyi A, Havass Z, Jansen KP, Wolf B. Neonatal screening for biotinidase deficiency in Hungary: clinical, biochemical and molecular studies. J Inherit Metab Dis. 2003;26:693–698. doi: 10.1023/b:boli.0000005622.89660.59. [DOI] [PubMed] [Google Scholar]
- 36.Neto EC, Schulte J, Rubim R, Lewis E, DeMari J, Castilhos C, Brites A, Giugliani R, Jensen KP, Wolf B. Newborn screening for biotinidase deficiency in Brazil: biochemical and molecular characterizations. Braz J Med Biol Res. 2004;37:295–299. doi: 10.1590/s0100-879x2004000300001. [DOI] [PubMed] [Google Scholar]
- 37.Brenner C. Catalysis in the nitrilase superfamily. Curr Opin Struct Biol. 2002;12:775–782. doi: 10.1016/s0959-440x(02)00387-1. [DOI] [PubMed] [Google Scholar]
- 38.Maras B, Barra D, Dupre S, Pitari G. Is pantetheinase the actual identity of mouse and human vanin-1 proteins. FEBS Lett. 1999;461:149–152. doi: 10.1016/s0014-5793(99)01439-8. [DOI] [PubMed] [Google Scholar]


