Abstract
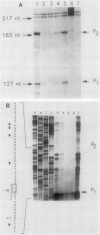
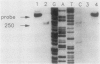
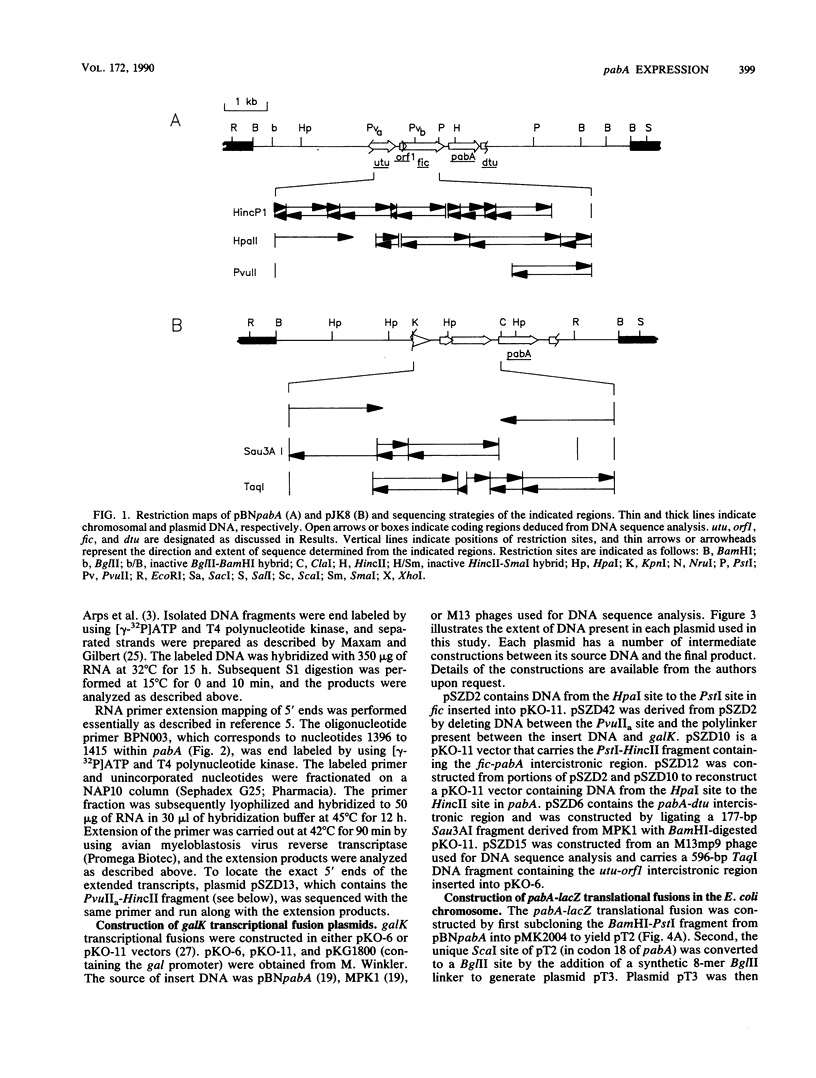
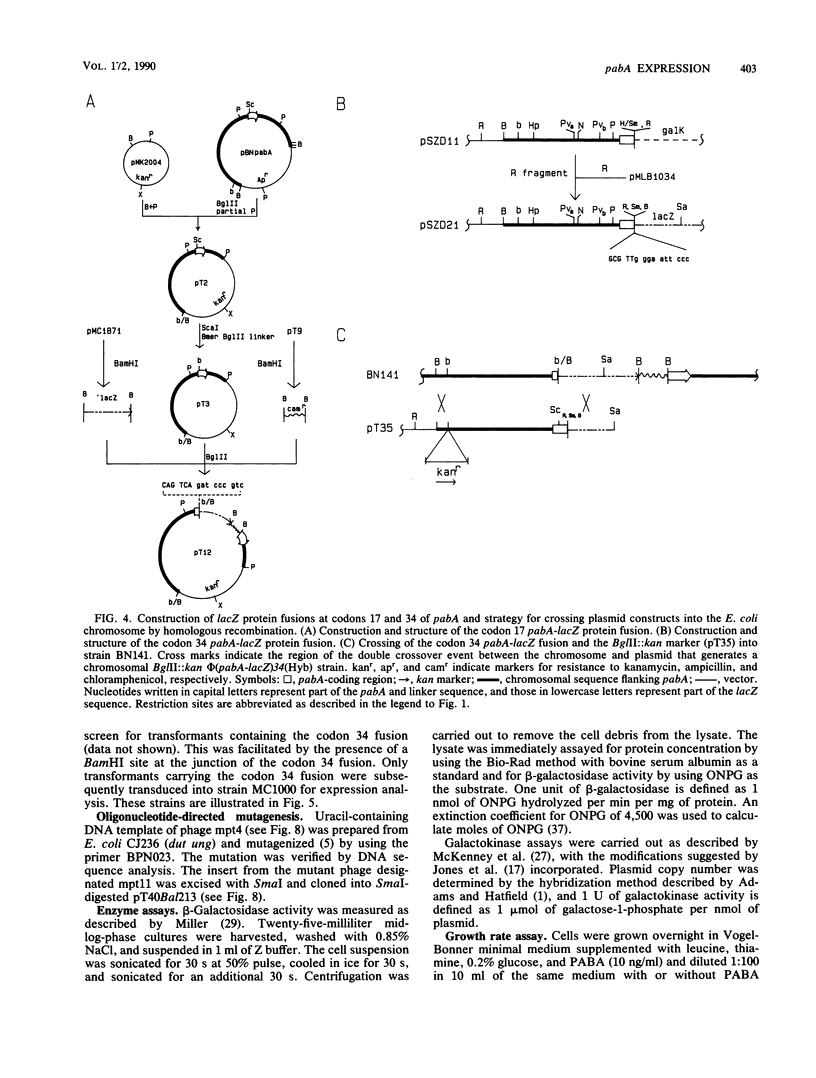
The pabA gene in Escherichia coli and Salmonella typhimurium encodes the glutamine amidotransferase subunit of para-aminobenzoate synthase, which catalyzes the first reaction in the conversion of chorismate to para-aminobenzoate (PABA). We have determined the nucleotide sequences of 1,362 base pairs preceding E. coli pabA and of 981 base pairs preceding S. typhimurium pabA. The nucleotide sequences suggest the presence of two protein-coding regions immediately upstream of pabA, designated orf1 and fic. Transcription analysis indicates that E. coli pabA is encoded by two overlapping transcriptional units. The polycistronic transcriptional unit includes orf1-fic-pabA and is initiated by the promoter designated P2. The monocistronic unit includes only pabA and is initiated by the promoter designated P1, which is located in the fic-coding region. Both promoters transcribe pabA to about the same steady-state level. However, expression analysis using chromosomal pabA-lacZ translational fusions indicated that P1 expressed PabA at least 50-fold more efficiently than P2. pabA-dependent growth rate analysis indicates that P1 is essential and P2 is dispensable for PABA metabolism. In the absence of P1, growth was reduced as a result of insufficient PabA expressed from P2. The significance of these results and possible posttranscriptional control mechanisms which affect PabA expression from the P2-initiated polycistronic unit are discussed.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. W., Hatfield G. W. Effects of promoter strengths and growth conditions on copy number of transcription-fusion vectors. J Biol Chem. 1984 Jun 25;259(12):7399–7403. [PubMed] [Google Scholar]
- Aksoy S., Squires C. L., Squires C. Translational coupling of the trpB and trpA genes in the Escherichia coli tryptophan operon. J Bacteriol. 1984 Feb;157(2):363–367. doi: 10.1128/jb.157.2.363-367.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arps P. J., Marvel C. C., Rubin B. C., Tolan D. A., Penhoet E. E., Winkler M. E. Structural features of the hisT operon of Escherichia coli K-12. Nucleic Acids Res. 1985 Jul 25;13(14):5297–5315. doi: 10.1093/nar/13.14.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arps P. J., Winkler M. E. Structural analysis of the Escherichia coli K-12 hisT operon by using a kanamycin resistance cassette. J Bacteriol. 1987 Mar;169(3):1061–1070. doi: 10.1128/jb.169.3.1061-1070.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bognar A. L., Osborne C., Shane B., Singer S. C., Ferone R. Folylpoly-gamma-glutamate synthetase-dihydrofolate synthetase. Cloning and high expression of the Escherichia coli folC gene and purification and properties of the gene product. J Biol Chem. 1985 May 10;260(9):5625–5630. [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Breeze A. S., Sims P., Stacey K. A. Trimethoprim-resistant mutants of E. coli K12: preliminary genetic mapping. Genet Res. 1975 Jun;25(3):207–214. doi: 10.1017/s0016672300015640. [DOI] [PubMed] [Google Scholar]
- Brown G. M., Williamson J. M. Biosynthesis of riboflavin, folic acid, thiamine, and pantothenic acid. Adv Enzymol Relat Areas Mol Biol. 1982;53:345–381. doi: 10.1002/9780470122983.ch9. [DOI] [PubMed] [Google Scholar]
- Cone K. C., Steege D. A. Functional analysis of lac repressor restart sites in translational initiation and reinitiation. J Mol Biol. 1985 Dec 20;186(4):733–742. doi: 10.1016/0022-2836(85)90393-6. [DOI] [PubMed] [Google Scholar]
- Goncharoff P., Nichols B. P. Nucleotide sequence of Escherichia coli pabB indicates a common evolutionary origin of p-aminobenzoate synthetase and anthranilate synthetase. J Bacteriol. 1984 Jul;159(1):57–62. doi: 10.1128/jb.159.1.57-62.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Gibson F. Biosynthesis of 4-aminobenzoate in Escherichia coli. J Bacteriol. 1970 Jun;102(3):767–773. doi: 10.1128/jb.102.3.767-773.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Pittard J. Genetic analysis of mutant strains of Escherichia coli requiring p-aminobenzoic acid for growth. J Bacteriol. 1967 Jun;93(6):1938–1942. doi: 10.1128/jb.93.6.1938-1942.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Jones H. M., Brajkovich C. M., Gunsalus R. P. In vivo 5' terminus and length of the mRNA for the proton-translocating ATPase (unc) operon of Escherichia coli. J Bacteriol. 1983 Sep;155(3):1279–1287. doi: 10.1128/jb.155.3.1279-1287.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J. B., Merkel W. K., Nichols B. P. Evolution of glutamine amidotransferase genes. Nucleotide sequences of the pabA genes from Salmonella typhimurium, Klebsiella aerogenes and Serratia marcescens. J Mol Biol. 1985 Jun 5;183(3):327–340. doi: 10.1016/0022-2836(85)90004-x. [DOI] [PubMed] [Google Scholar]
- Kaplan J. B., Nichols B. P. Nucleotide sequence of Escherichia coli pabA and its evolutionary relationship to trp(G)D. J Mol Biol. 1983 Aug 15;168(3):451–468. doi: 10.1016/s0022-2836(83)80295-2. [DOI] [PubMed] [Google Scholar]
- Kawamukai M., Matsuda H., Fujii W., Nishida T., Izumoto Y., Himeno M., Utsumi R., Komano T. Cloning of the fic-1 gene involved in cell filamentation induced by cyclic AMP and construction of a delta fic Escherichia coli strain. J Bacteriol. 1988 Sep;170(9):3864–3869. doi: 10.1128/jb.170.9.3864-3869.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamukai M., Matsuda H., Fujii W., Utsumi R., Komano T. Nucleotide sequences of fic and fic-1 genes involved in cell filamentation induced by cyclic AMP in Escherichia coli. J Bacteriol. 1989 Aug;171(8):4525–4529. doi: 10.1128/jb.171.8.4525-4529.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looman A. C., Bodlaender J., de Gruyter M., Vogelaar A., van Knippenberg P. H. Secondary structure as primary determinant of the efficiency of ribosomal binding sites in Escherichia coli. Nucleic Acids Res. 1986 Jul 11;14(13):5481–5497. doi: 10.1093/nar/14.13.5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looman A. C., van Knippenberg P. H. Effects of GUG and AUG initiation codons on the expression of lacZ in Escherichia coli. FEBS Lett. 1986 Mar 3;197(1-2):315–320. doi: 10.1016/0014-5793(86)80349-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCarthy J. E., Schauder B., Ziemke P. Post-transcriptional control in Escherichia coli: translation and degradation of the atp operon mRNA. Gene. 1988 Dec 10;72(1-2):131–139. doi: 10.1016/0378-1119(88)90135-7. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Nichols B. P., Seibold A. M., Doktor S. Z. para-aminobenzoate synthesis from chorismate occurs in two steps. J Biol Chem. 1989 May 25;264(15):8597–8601. [PubMed] [Google Scholar]
- Oppenheim D. S., Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980 Aug;95(4):785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P., Peterkofsky A., McKenney K. Translational efficiency of the Escherichia coli adenylate cyclase gene: mutating the UUG initiation codon to GUG or AUG results in increased gene expression. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5656–5660. doi: 10.1073/pnas.82.17.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K., Beck C. F. Point mutations that affect translation initiation of the transposon Tn10 tetA gene. Gene. 1988 Dec 30;74(2):559–563. doi: 10.1016/0378-1119(88)90190-4. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. Codon usage in regulatory genes in Escherichia coli does not reflect selection for 'rare' codons. Nucleic Acids Res. 1986 Oct 10;14(19):7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon R. Altered dihydrofolate reductase in fol regulatroy mutants of Escherichia coli K12. Mol Gen Genet. 1977 Mar 7;151(2):215–219. doi: 10.1007/BF00338697. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. E., Straus D. B., Grossman A. D., Burton Z. F., Gross C. A., Burgess R. R. Transcription from a heat-inducible promoter causes heat shock regulation of the sigma subunit of E. coli RNA polymerase. Cell. 1984 Sep;38(2):371–381. doi: 10.1016/0092-8674(84)90492-6. [DOI] [PubMed] [Google Scholar]
- Thomas L. K., Dix D. B., Thompson R. C. Codon choice and gene expression: synonymous codons differ in their ability to direct aminoacylated-transfer RNA binding to ribosomes in vitro. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4242–4246. doi: 10.1073/pnas.85.12.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Weinstock G. M., Berman M. L., Silhavy T. J. Chimeric genetics with beta-galactosidase. Gene Amplif Anal. 1983;3:27–64. [PubMed] [Google Scholar]
- White B. J., Hochhauser S. J., Cintron N. M., Weiss B. Genetic mapping of xthA, the structural gene for exonuclease III in Escherichia coli K-12. J Bacteriol. 1976 Jun;126(3):1082–1088. doi: 10.1128/jb.126.3.1082-1088.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Elledge S. J., Krueger J. H., Walker G. C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985 Mar;161(3):1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]