Abstract
In Drosophila, the fat body undergoes a massive burst of autophagy at the end of larval development in preparation for the pupal transition. To identify genes involved in this process, we carried out a microarray analysis. We found that mRNA levels of the homologs of Atg8, the coat protein of early autophagic structures, and lysosomal hydrolases were upregulated, consistent with previous results. Genes encoding mitochondrial proteins and many chaperones were downregulated, including the inhibitor of eIF2alpha kinases and the peptidyl-prolyl cis-trans isomerase (PPiase) FKBP39. Genetic manipulation of FKBP39 expression had a significant effect on autophagy, potentially through modulation of the transcription factor Foxo. Accordingly, we found that Foxo mutants can not properly undergo autophagy in response to starvation, and that overexpression of Foxo induces autophagy.
Keywords: autophagy, Drosophila, FKBP39, Foxo, microarray
Autophagy is the degradation of self material by lysosomes. As a primary cellular defense response, it is activated by nitrogen or amino acid starvation from yeast to mammals, and promotes the survival of the cell or organism by recycling dispensable cellular constituents for re-use in synthetic processes. The morphology of the major pathway, macroautophagy (referred to as ‘autophagy’ hereafter) is well-known for decades by electron microscopical studies (1). In response to starvation or other stimuli, a membranous sac called the phagophore or isolation membrane forms and engulfs portions of the cytosol. After sealing of its edges, the emerging double-membrane organelle is referred to as an autophagosome or initial autophagic vacuole (AVi). It subsequently fuses with a lysosome, resulting in the formation of an autolysosome or degrading autophagic vacuole (AVd), where degradation of the sequestered cellular material takes place. The process of autophagy is remarkably similar in all eukaryotic organisms, which suggests the involvement of an evolutionary conserved set of genes. Indeed, functional homologs of most Atg (autophagy-related) genes required for autophagy in yeast can also be found in multiple species including plants, worms, flies, and mammals (2). Despite the conservation of the core mechanism, there must be changes in the regulation of autophagy among different phyla, as it is involved in various cellular processes in multicellular animals. In addition to its fundamental role in starvation survival, autophagy is thought to play a role in cell death, neurodegeneration diseases, aging, immunity, growth, and cancer (for details, please consult recent reviews (1, 3, 4)).
In Drosophila and other insects undergoing complete metamorphosis, a tissue known as fat body acts as a store of proteins and other materials, which are released through autophagy to provide energy and nutrients during metamorphosis and early adulthood (5-8). The fat body, an analogue of the human liver, is a polytenic tissue that grows in mass approximately 200-fold in feeding Drosophila larvae during the three larval stages. After reaching an optimal mass, mature larvae stop eating and wander away from the food to find a suitable place for pupariation. At this time, the fat body undergoes a massive induction of autophagy, referred to below as developmental autophagy. These changes are induced by the insect molting hormone ecdysone at a low concentration of juvenile hormone (9). Recent results showed that ecdysone induces autophagy through downregulation of phosphatidyl-inositol 3-kinase (PI3K) signaling (10).
A central regulator of cell growth and autophagy is Tor (target of rapamycin) kinase. Inhibition of Tor activity rapidly results in growth arrest and induction of autophagy, which probably involves multiple phosphorylation and dephosphorylation events (11-13). In yeast, the phosphorylation state of a number of Atg proteins is rapamycin sensitive, and the activity of the kinase Atg1 is regulated by Tor signaling (14). Another potential regulatory mechanism is the induction of genes necessary for autophagy, or repression of genes that normally inhibit the process. It is known that the gene encoding Atg8, a ubiquitin-like coat protein for early autophagic structures, is upregulated in starved yeast cells (15). The mRNA level of one of its Drosophila homologs, CG32672/Atg8a (formerly known as CG1534), but not of other Atg gene homologs, was also shown to increase in response to starvation (16).
To search for genes regulated during developmental autophagy, we carried out a microarray analysis by comparing the transcriptional profiles of fat bodies dissected from feeding and wandering third instar larvae. This analysis both demonstrated evolutionary conservation and identified additional genes with previously unknown roles in autophagy. Further characterization of a selected subset of genes in transgenic animals identified FKBP39 as an inhibitor of autophagy, which effect is likely mediated through modulation of the transcription factor Foxo.
Results
1. Microarray analysis of transcriptional changes during developmental autophagy
To assess gene expression changes during developmental autophagy in larval Drosophila fat body, we manually dissected fat bodies before and after the developmental induction of autophagy from feeding (approximately 60 hours after hatching from the egg, Figure 1a) and wandering (84 hours after hatching from the egg, Figure 1b, e) third instar larvae (6, 7). Samples were processed and cDNAs were hybridized to a microarray containing 3200 annotated Drosophila cDNAs (17). 1941 of the 3200 genes investigated were expressed in the fat body. Table 1 shows the 57 genes induced by 1.65-fold or greater (estimated p-value <0.025) during autophagy. The mRNA level of the eye pigment biosynthesis gene Hn was increased, consistent with the known role of the fat body in synthesis of eye pigments during the wandering stage (18). A gene encoding Fbp2, a storage protein, was also induced; together these results provide a control for the proper developmental timing of our sample collection. Genes encoding putative lysosomal hydrolases (alpha-EST2, cathD, CG5932, CG1827, CG10992, CG1774) were upregulated, consistent with the expansion of the lysosomal compartment during autophagy seen by Lysotracker staining. 10 of the 16 fruit fly Atg gene homologs were represented on our chip, and only CG32672/Atg8a was induced significantly, in accordance with yeast and previous fruit fly data (1, 16). A gene encoding another ubiquitin-like protein of unknown function (CG7224) was also upregulated.
Figure 1.
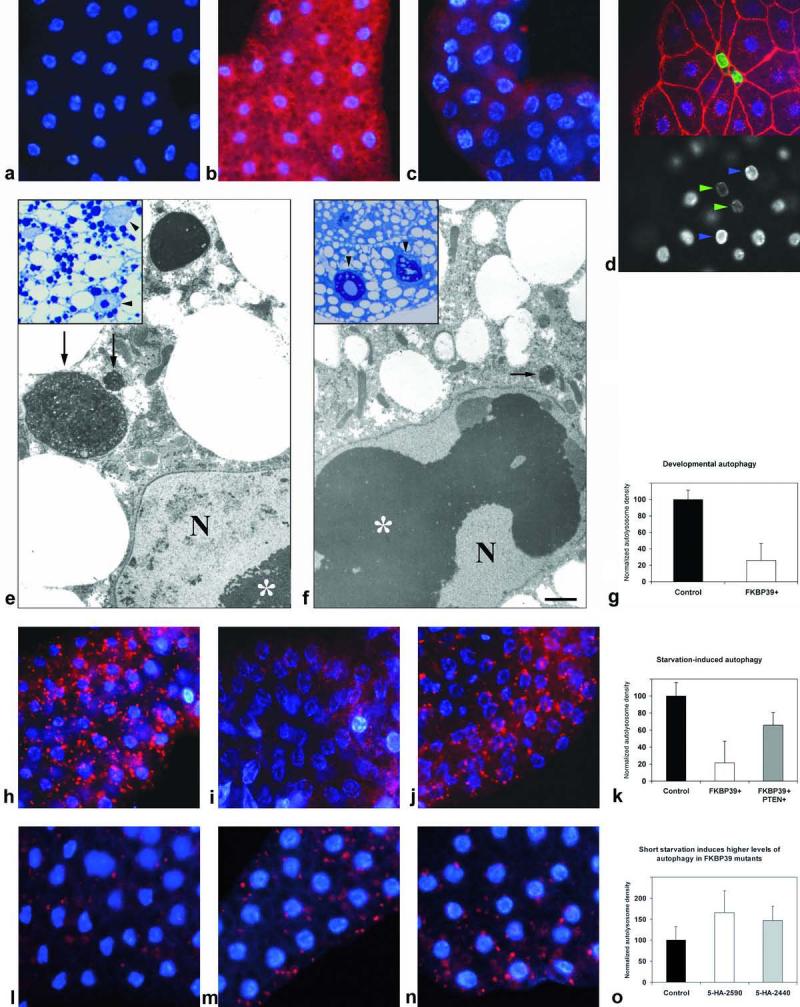
Overexpression of FKBP39 inhibits developmental and starvation-induced autophagy, whereas loss of FKBP39 function leads to higher than wild-type induction of autophagy.
a-c: Lysotracker staining of fat bodies. No Lysotracker staining is seen in feeding early third instar larvae (a), whereas Lysotracker-positive granules (shown in red) accumulate in fat body cells of wandering late third instars (b). Overexpression of FKBP39 in the fat body inhibits the formation of Lysotracker-positive granules (c), and leads to an inhibition of cell growth. This effect is best seen by cortical actin staining (red) of fat bodies clonally overexpressing FKBP39, marked by co-overexpression of nuclear GFP (green) (d, top panel). Lower panel shows DNA staining. Note that the nuclei of FKBP39-overexpressing cells (green arrows) are of the same size but contain less DNA than surrounding wild-type cells (blue arrows). e-f: electron microscopic images show fat body cells of late third instar larvae. Developmental autophagy results in the accumulation of large autolysosomes in wild-type larvae (arrow in e), whereas only small autolysosomes are seen in FKBP39-overexpressing fat body cells of the same age (arrows in f). Note the greatly enlarged nucleolus in f (asterisk), also caused by the overexpression of FKBP39. g shows quantitation of Lysotracker data on developmental autophagy. h-j: Lysotracker staining of fat bodies. Autophagy in fat body cells of early third instar larvae, induced by a 4-hour starvation (h, compare to a). Fat body-specific overexpression of FKBP39 also blocks this starvation response (i). Co-overexpression of PTEN restores autophagy inhibited by FKBP39 (j). k shows quantitation of Lysotracker data. l-n: Lysotracker staining of fat bodies. Short (80-minute) starvation induces higher levels of autophagy in fat bodies of FKBP39 mutant larvae than in wild-type controls (m, n, compare to l). o shows quantification of the results.
Panels a-d, h-j and l-n are of the same magnification (200x). Panels e-f are of the same magnification and the bar equals 1 um. N marks the nucleus in electron microscopical images. Error bars represent standard deviation in panels g, k and o. Genotypes: UASFKBP39/+ (a, b, e, h), cgGal4/UASFKBP39 (c, f, i), hsFLP; UASFKBP39/+; Act>CD2>Gal4, UASGFPnls/+ (d), cgGal4/UASFKBP39, UASPTEN (j), w1118 (l), w1118; 5-HA-2590/Df(3R)Exel6194 (m), w1118; 5-HA-2440/Df(3R)Exel6194 (n).
Table 1.
Genes upregulated during developmental autophagy in the Drosophila larval fat body.
Table shows the genes upregulated by at least 1.65-fold during developmental autophagy, with an estimated p-value of <0.025. In the first column, gene names are listed as in Flybase (21); second column shows fold induction relative to early third instars (prior to the onset of autophagy); third column shows the function, family or known domains of the genes listed.
| Gene name | Fold change | Function/Family/Domains | p-value |
|---|---|---|---|
| Cyp9f2 | 3.95 | oxidoreductase (cytochrome P450) | <0.0001 |
| cutlet | 3.54 | serine-type endopeptidase | <0.0001 |
| Fbp2 | 3.08 | nutrient reservoir; oxidoreductase | <0.0001 |
| alpha-Est2 | 2.77 | carboxylesterase | <0.0001 |
| CG7224 | 2.73 | ubiquitin-like | <0.0001 |
| Gs1 | 2.53 | glutamate synthase | 0.0002 |
| CG12262 | 2.52 | acyl-CoA dehydrogenase | 0.0002 |
| cathD | 2.29 | cathepsin D | 0.0007 |
| Reg-2 | 2.28 | haloacid dehydrogenase-like hydrolase | 0.0007 |
| CG9431 | 2.16 | immunoglobulin-like | 0.0014 |
| CG6440 | 2.16 | neuropeptide hormone | 0.0014 |
| syt | 2.16 | calcium-dependent phospholipid binding | 0.0015 |
| CG18418 | 2.14 | membrane carrier | 0.0016 |
| CG6957 | 2.14 | glucoseamine-6-phosphate deaminase | 0.0017 |
| Cyp4d2 | 2.12 | oxidoreductase (cytochrome P450) | 0.0018 |
| CG7523 | 2.12 | unknown | 0.0018 |
| l(2)k09913 | 2.11 | unknown | 0.0019 |
| Hn | 2.06 | eye pigment biosynthesis | 0.0026 |
| CG15309 | 2.06 | Yippee-like protein | 0.0026 |
| CG12428 | 2.02 | acetyltransferase | 0.0034 |
| CG5932 | 1.99 | triacylglycerol lipase | 0.0039 |
| cry | 1.97 | G-protein coupled photoreceptor activity | 0.0045 |
| CG1827 | 1.97 | aminohydrolase | 0.0046 |
| CG13603 | 1.94 | unknown | 0.0053 |
| CG30053 | 1.91 | unknown | 0.0061 |
| CG12323 | 1.91 | endopeptidase (proteasome b5 subunit) | 0.0061 |
| CG9813 | 1.90 | unknown | 0.0065 |
| CG7221 | 1.90 | oxidoreductase | 0.0066 |
| CG18787 | 1.89 | unknown | 0.0069 |
| CG4600 | 1.89 | acetyl-CoA acyltransferase | 0.0071 |
| Paip2 | 1.88 | translational repression | 0.0073 |
| CG8252 | 1.88 | unknown | 0.0075 |
| Atg8a | 1.86 | coat protein for autophagosomes | 0.0083 |
| CG2767 | 1.86 | alcohol dehydrogenase | 0.0084 |
| Keren | 1.84 | epidermal growth factor (EGF) | 0.0094 |
| CG12292 | 1.84 | unknown | 0.0095 |
| CG5738 | 1.84 | RNA polymerase II transcription factor | 0.0097 |
| Pdh | 1.83 | photoreceptor dehydrogenase | 0.0101 |
| CG17597 | 1.82 | acyl-CoA C-acyltransferase | 0.0108 |
| CG4669 | 1.77 | unknown | 0.014 |
| CG10007 | 1.77 | unknown | 0.0144 |
| CG1236 | 1.76 | phosphoglycerate dehydrogenase | 0.0149 |
| CG9286 | 1.76 | unknown | 0.015 |
| Trn-SR | 1.74 | nuclear import | 0.0163 |
| FucT6 | 1.73 | fucosyltransferase | 0.0178 |
| CG10992 | 1.73 | cathepsin B | 0.0179 |
| CG7789 | 1.72 | phosphatidylinositol phosphatase | 0.0182 |
| CG10433 | 1.72 | defense response | 0.0187 |
| CG32698 | 1.71 | carbonate dehydratase | 0.0191 |
| CG12428 | 1.71 | acetyltransferase | 0.0194 |
| CG8449 | 1.71 | unknown | 0.0196 |
| Ote | 1.71 | lamino-associated polypeptide 2 | 0.02 |
| CG14629 | 1.70 | unknown | 0.0202 |
| fng | 1.70 | glycosyltransferase | 0.0202 |
| CG1774 | 1.70 | lipid hydrolase | 0.0206 |
| CG12063 | 1.69 | Endoglin/CD105 antigen, PAN domains | 0.0215 |
| CG5721 | 1.68 | unknown | 0.023 |
The expression of 39 genes was significantly downregulated. Among them, two main subgroups could be identified: genes encoding mitochondrial proteins (CG17896, CG9140, ND42, CG6459, TRAP1, CG10664, Hsc70-5, CG2249, mRpL24, Marf) and cellular chaperones (CG8286/dIPK, CG4164, TRAP1, Hsc70-5, FKBP39). The expression level of Baldspot, encoding a transmembrane protein, also strongly decreased (Table 2).
Table 2.
Genes downregulated during developmental autophagy in the Drosophila larval fat body.
The list of genes downregulated by at least 1.65-fold during developmental autophagy with an estimated p-value of <0.025 (see legend of Table 1 for details).
| Gene name | Fold change | Function/Family/Domains | p-value |
|---|---|---|---|
| Baldspot | −4.69 | transmembrane protein | <0.0001 |
| CG17896 | −2.73 | methylmalonate-semialdehyde dehydrogenase | <0.0001 |
| CG3835 | −2.71 | oxidoreductase | <0.0001 |
| CG14683 | −2.56 | methyltransferase | 0.0001 |
| CG5171 | −2.37 | trehalose-phosphatase | 0.0003 |
| CG9140 | −2.37 | NADH dehydrogenase (ubiquinone) | 0.0003 |
| Nop56 | −2.15 | small nuclear ribonucleoprotein complex subunit | 0.0011 |
| CG8286 | −2.15 | Drosophila inhibitor of protein kinases (dIPK) | 0.0011 |
| CG4164 | −2.10 | Hsp40 | 0.0015 |
| CG10340 | −2.08 | possibly involved in protein complex assembly | 0.0017 |
| ND42 | −2.08 | NADH dehydrogenase (ubiquinone) | 0.0017 |
| CG6459 | −2.00 | mitochondrial glycoprotein | 0.0027 |
| CG3136 | −1.97 | basic leucine zipper (bZIP) transcription factor | 0.0033 |
| TRAP1 | −1.95 | mitochondrial Hsp90 | 0.0038 |
| CG6164 | −1.92 | unknown | 0.0044 |
| CG9249 | −1.92 | methyltransferase | 0.0045 |
| CG18316 | −1.89 | unknown | 0.0055 |
| CG10664 | −1.89 | cytochrome-c oxidase | 0.0055 |
| Ate1 | −1.87 | arginyltransferase | 0.0062 |
| yip7 | −1.86 | serine-type endopeptidase | 0.0065 |
| CG8732 | −1.85 | acetate-CoA ligase | 0.0069 |
| Hsc70-5 | −1.82 | mitochondrial Hsp70 | 0.0083 |
| CG15771 | −1.81 | unknown | 0.0087 |
| CG11597 | −1.80 | protein serine/threonine phosphatase (PP2A-like) | 0.0092 |
| CG3358 | −1.78 | nuclease | 0.0104 |
| CG2249 | −1.77 | cytochrome-c oxidase | 0.0107 |
| Tm1 | −1.76 | tropomyosin | 0.0114 |
| Sc2 | −1.75 | unknown | 0.0123 |
| CG17278 | −1.74 | protease inhibitor | 0.0126 |
| CG8664 | −1.73 | unknown | 0.0137 |
| Jon65Aiii | −1.71 | serine-type endopeptidase | 0.0151 |
| mRpL24 | −1.69 | mitochondrial large ribosomal subunit | 0.017 |
| Marf | −1.69 | GTPase and dynamin domains | 0.0172 |
| CG3902 | −1.69 | acyl-CoA dehydrogenase | 0.0175 |
| Cyp310a1 | −1.68 | oxidoreductase (cytochrome p450) | 0.0186 |
| His2Av | −1.68 | DNA binding (histone) | 0.0187 |
| eIF-4G | −1.67 | translation initiation factor | 0.0198 |
| FKBP39 | −1.66 | peptidyl-prolyl cis-trans isomerase (PPiase) | 0.0208 |
2. Quantitation of expressional changes for a selected subset of genes
A subset of genes potentially involved in autophagy were selected for further analysis by Quantitative Real-Time PCR (QRT-PCR) to confirm and more precisely quantitate the changes in mRNA expression levels. In addition we also analyzed expression of a few genes not represented on our chip, including Tor, CG12334/Atg8b, and CG15283, which encodes a ubiquitin-like protein highly similar to CG7224.
As shown in Table 3, these data well correlated with the results of the microarray analysis, and further confirmed the strong upregulation of both Atg8 homologs (CG32672/Atg8a: 6,96x and CG12334/Atg8b: 3,53x) and cathepsin D (CG1548: 2,14x), a well-known lysosomal hydrolase. The two ubiquitin-related genes we tested were also induced (CG7224: 3,73x and CG15283: 4,62x). Tor kinase mRNA level did not change, whereas genes encoding the cellular chaperones FKBP39 (FK506-binding protein of 39 kDa (19)) and CG8286/dIPK (Drosophila inhibitor of protein kinases (20)), and the transmembrane protein Baldspot (21) were strongly repressed (FKBP39: 6,73x, CG8286/dIPK: 4,92x, and Baldspot: 9,91x repression).
Table 3.
Quantitative Real-Time PCR analysis of the expression of a selected set of genes during developmental autophagy.
QRT-PCR analysis was used to confirm and further quantitate the expression changes seen in the microarray experiment, and also to analyze genes not physically represented on our chip. Gene names are listed in the first column; second column shows fold change based on the microarray experiment; third column shows fold change based on QRT-PCR. See text for details.
| Gene | Microarray | QRT-PCR |
|---|---|---|
| Atg8a | 1.86 | 6.96 |
| CG15283 | not on chip | 4.62 |
| CG7224 | 2.73 | 3.73 |
| Atg8b | not on chip | 3.53 |
| cathD | 2.29 | 2.14 |
| Tor | not on chip | no change |
| CG8286/dIPK | −2.15 | −4.92 |
| FKBP39 | −1.66 | −6.73 |
| Baldspot | −4.69 | −9.91 |
3. Analysis of candidate autophagy-related genes in transgenic animals identified FKBP39 as an inhibitor of autophagy
To establish the role of selected genes regulated in autophagy, we carried out overexpression studies using the Gal4-UAS system. Overexpression of most genes we tested produced various phenotypes with different Gal4 drivers, but had no effect on autophagy (Supplementary Table 1). For example, expression of cathD, Atg8a, Baldspot, and dIPK neither induced nor inhibited developmental autophagy when expressed in the fat body or throughout the whole animal.
In contrast to the above results, fat body-specific overexpression of FKBP39, a peptidyl-prolyl cis-trans isomerase (PPiase) had a dramatic effect on developmental autophagy. Very few autolysosomes were observed in fat body cells of wandering larvae overexpressing FKBP39, and these were also smaller than in wild-type animals (Figure 1c, f, compare to Figure 1b, e; see also Figure 1g for quantitation of the results). Interestingly, FKBP39 overexpression also inhibited cell growth. The average size of fat body cells overexpressing FKBP39 was 43±21% of wild-type neighboring cells (Figure 1d). The overexpressing cells also had a lower DNA content, suggesting a delay in endoreduplication. The fact that the nuclei were about the same size as the nuclei of wild-type cells is probably due to the greatly enlarged nucleolus (Figure 1d, see also Figure 1f).
We next tested if overexpression of FKBP39 had an effect on starvation-induced autophagy. Starving early third instar larvae for 4 hours in sucrose solution results in strong accumulation of Lysotracker-positive autolysosomes in larval fat body cells (13)(Figure 1h, compare to Figure 1a). Overexpression of FKBP39 in fat body cells greatly inhibited the induction of autophagy in starved animals (Figure 1i; see also Figure 1k for quantitation of the results), similar to the case of developmental autophagy.
As a final verification, we then tested how loss of FKBP39 function influences autophagy using the mutant lines 5-HA-2590 and 5-HA-2440. These hypomorphic mutants harbor P-element insertions in the 5' non-translated region of FKBP39. Although we were unable to observe ectopic induction of autophagy in these mutants (not shown), we found that these larvae displayed a significantly higher level of autophagy than wild-type controls in response to a short, 80-minute starvation (Figure 1l-n, see Figure 1o for quantification of the results; p=0.001 for 5-HA-2590/Df(3R)Exel6174, and p=0.006 for 5-HA-2440/Df(3R)Exel6174).
4. Co-overexpression of PTEN restores autophagy inhibited by FKBP39
To identify the signal transduction pathway FKBP39 may act through, we tested several signaling cascades that were previously reported to be influenced by FKBPs (22, 23). Modulation of Ras (by overexpressing wild-type, hyperactive or dominant-negative Ras or wild-type Raf), protein kinase A (by overexpressing wild-type PKAc or its inhibitors PKI or mutant PKAr), or S6 kinase (by overexpressing hyperactive S6K) signaling had no effect on developmental autophagy, and did not restore autophagy in FKBP39-overexpressing fat bodies, as analyzed by Lysotracker staining (not shown). As Ras signaling was suggested to be involved in the induction of autophagy in other systems, and FKBP39 inhibited the Ras-mediated activation of MAPK (Supplementary Figure 1a), we analyzed autophagy by generating loss of function somatic clones of a Ras null mutation in the fat body. Although slightly smaller than surrounding wild-type cells, these cells showed normal induction of developmental autophagy (Supplementary Figure 1b), proving that Ras itself is not required for autophagy in the fly fat body.
In contrast to the above results, overexpression of PTEN, a phosphatase that antagonizes class I PI3K signaling, restored starvation-induced autophagy in FKBP39-expressing fat body cells (Figure 1j, see also Figure 1k for quantitation of the results). Unfortunately, we could not test developmental autophagy in this case, as simultaneous expression of these two genes in the fat body led to a prolonged third instar stage and ultimately lethality, potentially due to the cumulative growth-inhibiting effects of the two proteins.
5. The autophagy-inhibiting effect of FKBP39 is likely mediated by inhibition of Foxo, a novel regulator of autophagy
The above results suggested that FKBP39 may inhibit autophagy through activation of PI3K, a potent inhibitor of both starvation-induced and developmental autophagy (10, 13). To test this, we analyzed the level of PI3K signaling in FKBP39-overexpressing clones. GFP-pleckstrin homology (PH) domain fusion proteins are well-established markers of PI3K activity (24). In cells with high levels of PI3K activity, accumulation of the membrane lipid phosphatidylinositol-3,4,5-phosphate results in localization of the GFP-PH marker to the cell membrane. We found that FKBP39 overexpression led to a reduction of membrane versus cytoplasmic ratio of the signal compared to surrounding wild-type cells (Figure 2a). This result indicates that PI3K activity is reduced in FKBP39-overexpressing cells, thus discounting the possibility that FKBP39 inhibits autophagy by upregulating PI3K signaling.
Figure 2.
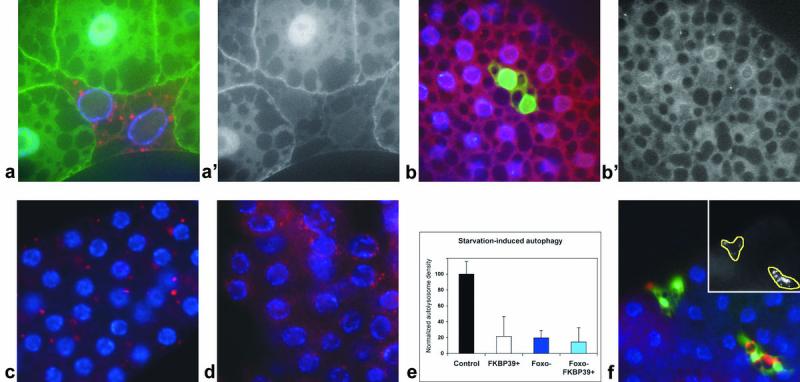
The autophagy-inhibiting effect of FKBP39 overexpression is potentially mediated by inhibition of Foxo.
a: the membrane localization of a probe used as an indicator of PI3K activity (GFP-PH, green) is greatly reduced compared to the cytoplasmic signal in fat body cells of feeding larvae that clonally overexpress FKBP39 (marked by punctate myrRFP expression, red). a‘ shows the green channel separately. b: Foxo staining (red) in fat bodies of wild-type mid-third instar larvae shows both nuclear and cytoplasmic labeling. In cells overexpressing FKBP39 (and GFP, green), the nuclear pool of Foxo is completely abolished, and only cytoplasmic signal is seen. b‘ shows the red channel separately. c: null mutation of Foxo (Foxo21/Foxo25) strongly decreases starvation-induced autophagy in the larval fat body, as shown by Lysotracker staining (red) (compare to Figure 1h). d: overexpression of FKBP39 in a Foxo null mutant background results in a reduction similar to that seen in case of Foxo mutation or FKBP39 overexpression alone, also shown by quantitation of Lysotracker data in panel e. f: overexpression of an activated form of Foxo, Foxo™ strongly induces autophagy in fat body cells of feeding larvae. Inset shows the red channel (Lysotracker staining) in the overexpressing cells (delineated by a yellow line). Magnification is 300x for panel a and 200x for panels b-d and f. Error bars represent standard deviation in panel e. Genotypes: hsFLP; UASFKBP39/UASmyrRFP; Act>CD2>Gal4, tGPH/+ (a), hsFLP; UASFKBP39/+; Act>CD2>Gal4, UASGFPnls/+ (b), Foxo21/Foxo25 (c), cgGal4/UASFKBP39; Foxo21/Foxo25 (d), hsFLP; UASFoxo™/+; Act>CD2>Gal4, UASGFPnls/+ (f).
As another potential measure of PI3K signaling, we also examined the localization of the transcription factor Foxo in FKBP39 overexpressing clones. Elevated PI3K signaling results in phosphorylation of Foxo by the serine/threonine kinase Akt (25). In response to this phosphorylation event, Foxo changes its intracellular localization: the nuclear pool translocates to the cytoplasm (25). Wild-type mid-third instar fat body cells displayed a moderate level of nuclear Foxo. Overexpression of FKBP39 led to a nearly complete exclusion of Foxo from the nucleus (Figure 2b), despite the reduced level of PI3K signaling in these cells. This result suggests that the autophagy-inhibiting effect of FKBP39 may be mediated through inhibition of Foxo.
To test this hypothesis, we carried out overexpression of FKBP39 in a Foxo null mutant background. Null mutation of Foxo alone strongly interfered with starvation-induced autophagy, indicating for the first time a role for Foxo in this process (Figure 2c, compare to Figure 1h). Overexpression of FKBP39 in a Foxo null mutant background failed to further suppress autophagy compared to mutation of Foxo or overexpression of FKBP39 alone (Figure 2d, compare to Figures 2c and 1i, respectively; see also Figure 2e for quantitation of these results), suggesting that the autophagy-inhibiting effect of FKBP39 is at least partly mediated by inhibition of Foxo.
Finally, we also expressed an activated version of Foxo (Foxo™) (25) in fat body cells of feeding larvae. We found that Foxo™ expression was sufficient to strongly induce autophagy, further confirming the important role of Foxo in regulating autophagy (Figure 2f). Together, our results indicate that Foxo is necessary and sufficient for proper induction of autophagy, and they identify Foxo as a relevant target of FKBP39 action.
Discussion
We have used microarrays containing 3200 annotated Drosophila cDNAs to identify genes that are regulated at the time of developmental autophagy in the larval fat body. Upregulated genes whose protein products likely function in developmental autophagy included various lysosomal hydrolases like cathepsin D and cathepsin B, and the ubiquitin-like coat proteins for initial autophagic vacuoles, Atg8a and Atg8b. Expansion of the lysosomal system is usually observed during autophagy, and the upregulation of hydrolase genes is consistent with the demand for increased degrading capacity during massive induction of autophagy. Of the fruit fly homologs of yeast autophagy genes, Atg8a and Atg8b showed induction, similarly to starved yeast cells or Drosophila larvae (1, 16). Genes of two additional small ubiquitin-like proteins, CG7224 and CG15283, were also strongly upregulated. Although the potential role of these genes in autophagy requires further studies, it is interesting to note that they share a strongly conserved human homolog, LOC135154.
Genes encoding various mitochondrial proteins were downregulated, which is consistent with the observation that a large number of mitochondria are degraded during developmental autophagy (5-7). mRNA levels of many cellular chaperones also decreased, as reported earlier (see below).
Gene expression profiling analyses under a variety of experimental conditions or developmental transitions in which induction of autophagy is observed have been reported previously (16, 26, 27). Although it is impossible to isolate autophagy from the numerous other cellular processes with which cells respond to environmental or developmental conditions that also induce autophagy, careful comparison of our results and the results from these studies enabled us to identify the most promising candidate genes that may play a common role during autophagy. Of the genes we identified as being induced during developmental autophagy in the larval fat body, several were previously found to be also upregulated in response to sucrose or no-sugar starvation, conditions that provoke a strong autophagic response in most polyploid larval tissues. These included CG7224, CG15309, CG13603, PAIP2, Atg8a and CG10992/cathepsin B (16). Moreover, all but one of these genes were also identified as upregulated during salivary gland cell death that likely involves autophagy (26, 27). The expression of Atg8, the coat protein for autophagosomes, which was identified as a rate-limiting factor for the main pathway of autophagy in yeast (15), showed only a slight induction in these studies. It may suggest that apart from the main pathway of autophagy, other forms of lysosomal degradation are potentially involved in salivary gland cell death. In fact, studies of Drosophila null mutants for dark, the fly homolog of the apoptosis gene Apaf-1, suggest that autophagy is not affected in the mutant salivary glands that fail to undergo histolysis (28), similar to hypomorphic Hid mutants or to flies overexpressing the caspase inhibitor p35 (29).
The expression of the genes Nop56, CG8286/dIPK, CG4164/Hsp40, CG6459, Hsc70-5, CG3902, His2Av and FKBP39 was also reduced both during developmental autophagy and in response to sucrose or no-sugar starvation (16). Three of these, CG8286/dIPK, CG4164/Hsp40 and Hsc70-5 were also downregulated in dying salivary glands (26, 27).
In this work, we chose three downregulated genes for detailed analysis. Overexpression of two of these, dIPK, a cellular chaperone, and Baldspot, a transmembrane protein, did not inhibit autophagy in our tests. However, it is important to note that overexpression of Atg8a or cathD had no effect on autophagy either, despite their very well established role during autophagy. Indeed, the hypomorphic Baldspot mutation l(3)02281 results in darker than usual body color (21). As pigment granules are lysosome-related organelles, it may suggest that Baldspot normally inhibits the biogenesis of these pigment granules, and its mutation results in the generation of more granules.
CG8286/dIPK is a cellular chaperone containing a heat shock protein DnaJ domain, a tetratricopeptide repeat that mediates protein-protein interactions, and a protein prenyltransferase domain. The human orthologue of dIPK is the 58 kD inhibitor of protein kinases p58IPK, which strongly inhibits eIF2alpha protein kinases PKR and PERK (30, 31). Phosphorylation of eIF2alpha results in global translational repression. eIF2alpha protein kinases function in mediating various stress signals such as the presence of double-stranded RNA viruses, interferon gamma, endoplasmic reticulum stress or starvation (32, 33). p58IPK therefore promotes normal translational activity, and it was also shown to inhibit virus- or TNFalpha-induced cell death by both eIF2alpha kinase-dependent and independent mechanisms (34). The yeast eIF2alpha kinase GCN2 is required for starvation-induced autophagy, and PKR null or non-phosphorylatable eIF2alpha mutant murine embryonic fibroblasts are defective in virus-induced autophagy (35). In our system, overexpression of dIPK did not interfere with developmental autophagy, suggesting that eIF2alpha kinase signaling is not the major pathway functioning during this process in Drosophila. Co-overexpression of dIPK partially rescued the phenotypes caused by expression of the proapoptotic protein reaper or the caspase dronc (our unpublished data), proving the evolutionary conserved anti-apoptotic function of dIPK.
FKBP39 is a small FKBP that belongs to the family of PPiases with 21 family members in Drosophila (21). PPiases catalyse the isomerization of the peptide bond between a proline and a bulky residue, including phosphoserine or phosphothreonine. This motif is generated through the action of proline-directed kinases, the best characterized examples of which are the kinases downstream of Ras. FKBPs are also the intracellular receptors of the immunosuppressive drugs rapamycin and FK506. The FKBP12-rapamycin complex binds to Tor kinase and inhibits cell growth and induces autophagy. However, in the absence of drug, no interaction is observed between these proteins. Physiological roles of FKBPs are diverse and not fully characterized yet, but FKBP12 is known to bind to kinases such as transforming growth factor-beta receptor or epidermal growth factor receptor and the phosphatase calcineurin and inhibit their activity. FKBP12 also binds to ER-resident calcium channels, modulating calcium release regulation by protein kinase A and calcineurin (22).
We identified FKBP39 as a downregulated gene during developmental autophagy in Drosophila, suggesting that FKBP39 may be an inhibitor of autophagy. Indeed, overexpression of FKBP39 in transgenic animals led to a strong inhibition of both developmental and starvation-induced autophagy and cell growth in the larval fat body, whereas loss of FKBP39 function resulted in a higher than wild-type induction of autophagy in response to a short starvation.
PI3K signaling is a major regulator of cell growth and autophagy, and it was recently shown that during developmental reprogramming of the fat body preceding metamorphosis, ecdysone induces autophagy in part by downregulating PI3K signaling (10). In that study, expression of a dominant-negative ecdysone receptor inhibited developmental autophagy, and co-overexpression of PTEN, the phosphatase that antagonizes PI3K activity, reversed this effect. We identified FKBP39 as a potential physiological inhibitor of autophagy, its overexpression causing effects similar to inhibition of ecdysone signaling. Co-overexpression of PTEN also reversed the inhibitory effect of FKBP39 overexpression on autophagy. However, in a more direct test of PI3K activity, we found that overexpression of FKBP39 decreased the membrane localization of a probe that reflects PI3K activity. These results indicate that autophagy is inhibited in FKBP39-overexpressing cells despite a reduction in PI3K signaling. Further, they suggest that the small size of these cells may be due in part to decreased PI3K activity.
Interestingly, overexpression of FKBP39 resulted in nuclear exclusion of Foxo, suggesting that FKBP39 may inhibit autophagy through inhibition of Foxo. Foxo is a Forkhead family transcription factor activated by decreased PI3K or increased stress signaling, and has been shown to be required for growth inhibition, increased stress resistance, and lifespan extension provoked by modulation of these signaling pathways, respectively (36, 37). Mutation of Foxo caused a similar reduction of starvation-induced autophagy as overexpression of FKBP39, and overexpression of activated Foxo was sufficient to induce autophagy in fat bodies of feeding larvae, demonstrating that Foxo is indeed involved in the regulation of autophagy. Overexpression of FKBP39 in a Foxo mutant background did not significantly decrease autophagy compared to cells in which Foxo was mutated or FKBP39 was overexpressed alone (p-values >0.2). This result suggests that at least part of the effect of FKBP39 overexpression on autophagy is mediated by its inhibition of Foxo, as one would expect additive effects of autophagy inhibition in the case of independent signaling pathways. Other effects caused by overexpression of FKBP39 (enlarged nucleolus and larval-prepupal lethality) were not affected by the Foxo mutant background (see Figure 2d, and not shown).
Given the known targets of PPiases (see earlier), the effect of FKBP39 on Foxo localization is probably indirect. In Drosophila, Foxo localization and activity has been shown to be regulated through phosphorylation by PI3K/Akt and Jun N-terminal kinase (JNK) signaling (25, 37). Based on our results presented here, PI3K signaling is unlikely to mediate the effects of FKBP39 overexpression on Foxo localization, although it remains possible that FKBP39 may affect Akt independently of PI3K. Therefore, JNK signaling is a promising candidate for mediating at least some of the effects of FKBP39 overexpression (23), especially considering the strong effect of FKBP39 overexpression on kinases downstream of Ras (see Supplementary Figure 1a) that are closely related to JNK family kinases. This issue clearly warrants future studies.
In summary, we have identified numerous genes regulated during developmental autophagy in larval Drosophila fat body, and we have shown that FKBP39, a gene downregulated during the process, encodes a likely physiological inhibitor of autophagy. We have also identified Foxo as a novel regulator of autophagy, potentially mediating the inhibitory effect of FKBP39 on autophagy.
Materials and methods
Drosophila lines and methods
The following Drosophila lines were used in our study: EP362 (Atg8a), EP2151 (cathD) (Szeged Stock Center), UASmyrRFP, hsGal4, cgGal4, GMRGal4, actGal4, tubGal4, eyGal4, Df(3R)Exel6174 (Bloomington Stock Center), 5-HA-2590 and 5-HA-2440 (kindly provided by Gunther Reuter), FRT82B Rasc40b (kindly provided by Celeste Berg), hsFLP, Act>CD2>Gal4, UASGFPnls and Act>CD2>Gal4, tGPH (kindly provided by Bruce A. Edgar), Foxo25 and Foxo21 (kindly provided by Heinrich Jasper and Ernst Hafen), and UASFoxo™ (kindly provided by Marc Tatar). Fruit flies were maintained at room temperature (23-25 oC). Heat shocks were carried out by immersing vials in a 37 °C water bath for 1 hour. Gain of function clones in the fat body were spontaneously generated by the leaky expression of FLP recombinase in hsFLP; Act>CD2>Gal4, UASGFPnls/UASFKBP39 animals (24). We generated Ras null mutant clones in the fat body by heat-shocking 0-8 hour old embryos of the genotype hsFLP; cgGal4/+; FRT82B, UASGFP/FRT82B, Rasc40b. As expected, no Ras null mutant cells were recovered in the eyes of eyFLP; FRT82B, M(3)95A/FRT82B, Rasc40b animals.
Molecular cloning and establishment of transgenic lines
Full-length cDNAs encoding Baldspot, dIPK and FKBP39 (EST# GH11554, LD25575 and LD30817, kindly provided by Miklós Erdélyi) were cloned into the pUAST fly transformation vector using EcoRI and XhoI restriction sites. Resulting plasmids were sequenced and injected into fruit fly embryos using standard microinjection techniques. Multiple transgenic lines were established and analyzed for each construct with similar results.
Microarrays, probe preparations, hybridizations, scanning, data analysis.
Construction and use of microarrays were performed as described (38). Briefly, 3200 amplified cDNA inserts from Drosophila melanogaster were purified with MultiScreen-PCR plate (Millipore), resuspended in 50% dimethylsulfoxide/water, and arrayed on amino-silanized slides (Sigma-Aldrich) by using a MicroGrid Total Array System (BioRobotics) spotter with 16 pins with a 4×4 format. All clones were spotted in duplicate. Prior to hybridization, the slides were blocked in 1 × SSC, 0.2% SDS, 1% BSA for 30 min at 42 °C, washed with water and dried with high pressure air. Fat bodies were manually dissected from 150-200 carefully staged feeding and wandering animals. All tissues except the embedded gonad disks were dissected away prior to freezing fat bodies in liquid nitrogen. 15 ug total RNA from each sample was amplified by a linear antisense RNA amplification method, and labeled with Cy3 or Cy5 fluorescent dye during reverse transcription as described previously (39). Probes were mixed, reconstituted in 12 ul hybridization buffer (50 % formamide, 5× SSC, 0.1 % SDS, 100 mg/ml salmon sperm DNA) and applied onto the array after denaturation by heating for 1 min at 90°C. The slide was covered and incubated at 42°C for 20 hours in a humid hybridization chamber. After hybridization the arrays were washed by submersion and agitation for 10 min in 1 × SSC with 0.1% SDS, for 10 min in 0.1% × SSC with 0.1% SDS and for 10 min in 0.1 × SSC at room temperature, then rinsed briefly in deionized water and dried. Each array was scanned under a green laser (532 nm) (for Cy3 labeling) and under a red laser (660 nm) (for Cy5 labeling) by using a ScanArray Lite (GSI Lumonics) scanning confocal fluorescent scanner with 10 um resolution. Scanned output files were analyzed using the GenePix Pro 3.0 software (Axon Instruments Inc.). Each spot was defined by automatic positioning of a grid of circles over the image. The average and median pixel intensity ratios calculated from both channels and the local background of each spot were determined. An average expression ratio (MeaR, denotes the average of local background corrected pixel intensity ratios) was determined for each spot. Data analysis was done by the Significance Analysis of Microarrays (SAM) method and visualization of scatter images were performed with the Microsoft EXCEL software.
Real-time quantitative PCR
Relative quantitative reverse transcription-PCR (QRT-PCR) was performed on a RotorGene 2000 instrument (Corbett Research) with gene-specific primers and SybrGreen protocol to confirm the gene expression changes observed by using microarrays. 20 ug of total RNA from each pool was reverse transcribed in the presence of poly(dT) sequences in total volume of 20 ul. After dilution of the mix with 80 ul of water, 2 ul of this mix was used as template in the QRT-PCR. Relative expression ratios were normalized to actin. The PCR primers used in this study are available upon request. All PCRs were performed in triplicates.
Histology
Lysotracker Red staining of dissected larval fat bodies, cortical actin-phalloidin staining and electron microscopy was done as described previously (8, 10, 13). Anti-Foxo antibody (25) was used at a dilution of 1:300.
Western blots
Western blots were carried out as described in (8), using an antibody specific to diphosphorylated MAPK (Sigma) at a dilution of 1:1000.
Supplementary Material
Acknowledgements
We would like to thank Zsoltné Pálfia, Mariann Saródy and Emese Léder for the excellent technical assistance, our colleagues listed in the Methods section for providing reagents, and our anonymous reviewers for their helpful comments. This work was supported by NIH grant RO1 GM62509 provided to T.P.N. and by the Hungarian Ministry of Education grant MEDICHEM 1/047 NKFP provided to M.S.
Abbreviations
- dIPK
Drosophila inhibitor of protein kinases
- FKBP39
FK506-binding protein of 39 kDa
- JNK
Jun N-terminal kinase
- QRT-PCR
quantitative real-time polymerase chain reaction
- PI3K
phosphatidyl-inositol 3-kinase
- PPiase
peptidyl-prolyl cis-trans isomerase
- Tor
target of rapamycin
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5(4):539–45. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535–41. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 5.Sass M, Kovacs J. The effect of ecdysone on the fat body cells of the penultimate larvae of Mamestra brassicae. Cell Tissue Res. 1977;180(3):403–9. doi: 10.1007/BF00227604. [DOI] [PubMed] [Google Scholar]
- 6.Butterworth FM, Forrest EC. Ultrastructure of the preparative phase of cell death in the larval fat body of Drosophila melanogaster. Tissue Cell. 1984;16(2):237–50. doi: 10.1016/0040-8166(84)90047-8. [DOI] [PubMed] [Google Scholar]
- 7.Butterworth FM, Emerson L, Rasch EM. Maturation and degeneration of the fat body in the Drosophila larva and pupa as revealed by morphometric analysis. Tissue Cell. 1988;20(2):255–68. doi: 10.1016/0040-8166(88)90047-x. [DOI] [PubMed] [Google Scholar]
- 8.Juhasz G, Csikos G, Sinka R, Erdelyi M, Sass M. The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 2003;543(13):154–8. doi: 10.1016/s0014-5793(03)00431-9. [DOI] [PubMed] [Google Scholar]
- 9.Sass M, Kovacs J. Ecdysterone and an analogue of juvenile hormone on the autophagy in the cells of fat body of mamestra brassicae. Acta Biol Acad Sci Hung. 1975;26(34):189–96. [PubMed] [Google Scholar]
- 10.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7(2):179–92. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148(3):465–80. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270(5):2320–6. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 13.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7(2):167–78. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147(2):435–46. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. Embo J. 2002;21(22):6162–73. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szuperak M, Zvara A, Erdelyi M. Identification of germ plasm-enriched mRNAs in Drosophila melanogaster by the cDNA microarray technique. Gene Expr Patterns. 2005;5(5):717–23. doi: 10.1016/j.modgep.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Tearle R. Tissue specific effects of ommochrome pathway mutations in Drosophila melanogaster. Genet Res. 1991;57(3):257–66. doi: 10.1017/s0016672300029402. [DOI] [PubMed] [Google Scholar]
- 19.Theopold U, Dal Zotto L, Hultmark D. FKBP39, a Drosophila member of a family of proteins that bind the immunosuppressive drug FK506. Gene. 1995;156(2):247–51. doi: 10.1016/0378-1119(95)00019-3. [DOI] [PubMed] [Google Scholar]
- 20.Melville MW, Katze MG, Tan SL. P58IPK, a novel cochaperone containing tetratricopeptide repeats and a J-domain with oncogenic potential. Cell Mol Life Sci. 2000;57(2):311–22. doi: 10.1007/PL00000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drysdale RA, Crosby MA. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–5. doi: 10.1093/nar/gki046. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrar Y, Bellini C, Faure JD. FKBPs: at the crossroads of folding and transduction. Trends Plant Sci. 2001;6(9):426–31. doi: 10.1016/s1360-1385(01)02044-1. [DOI] [PubMed] [Google Scholar]
- 23.Klettner A, Baumgrass R, Zhang Y, Fischer G, Burger E, Herdegen T, Mielke K. The neuroprotective actions of FK506 binding protein ligands: neuronal survival is triggered by de novo RNA synthesis, but is independent of inhibition of JNK and calcineurin. Brain Res Mol Brain Res. 2001;97(1):21–31. doi: 10.1016/s0169-328x(01)00286-8. [DOI] [PubMed] [Google Scholar]
- 24.Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2(2):239–49. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- 25.Hwangbo DS, Gersham B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429(6991):562–6. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 26.Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol. 2003;13(4):350–7. doi: 10.1016/s0960-9822(03)00085-x. [DOI] [PubMed] [Google Scholar]
- 27.Gorski SM, Chittaranjan S, Pleasance ED, Freeman JD, Anderson CL, Varhol RJ, Coughlin SM, Zuyderduyn SD, Jones SJ, Marra MA. A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol. 2003;13(4):358–63. doi: 10.1016/s0960-9822(03)00082-4. [DOI] [PubMed] [Google Scholar]
- 28.Akdemir F, Farkas R, Chen P, Juhasz G, Medvedova L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophay occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006 doi: 10.1242/dev.02332. In press. [DOI] [PubMed] [Google Scholar]
- 29.Juhasz G, Sass M. Hid can induce, but is not required for autophagy in polyploid larval Drosophila tissues. Eur J Cell Biol. 2005;84(4):491–502. doi: 10.1016/j.ejcb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci U S A. 2002;99(25):15920–5. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan SL, Gale MJ, Jr., Katze MG. Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol Cell Biol. 1998;18(5):2431–43. doi: 10.1128/mcb.18.5.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pain VM. Translational control during amino acid starvation. Biochimie. 1994;76(8):718–28. doi: 10.1016/0300-9084(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 33.Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18(45):6112–20. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 34.Tang NM, Korth MJ, Gale M, Jr., Wambach M, Der SD, Bandyopadhyay SK, Williams BR, Katze MG. Inhibition of double-stranded RNA- and tumor necrosis factor alpha-mediated apoptosis by tetratricopeptide repeat protein and cochaperone P58(IPK) Mol Cell Biol. 1999;19(7):4757–65. doi: 10.1128/mcb.19.7.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talloczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A. 2002;99(1):190–5. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, Hafen E. The Drosophila Forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003;2(3):20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121(1):115–25. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 38.Puskas LG, Zvara A, Hackler L, Jr., Micsik T, van Hummelen P. Production of bulk amounts of universal RNA for DNA microarrays. Biotechniques. 2002;33(4):898–900. doi: 10.2144/02334mt03. 902, 904. [DOI] [PubMed] [Google Scholar]
- 39.Puskas LG, Zvara A, Hackler L, Jr., Van Hummelen P. RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques. 2002;32(6):1330–4. doi: 10.2144/02326mt04. 1336, 1338, 1340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


