Abstract
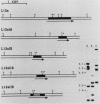
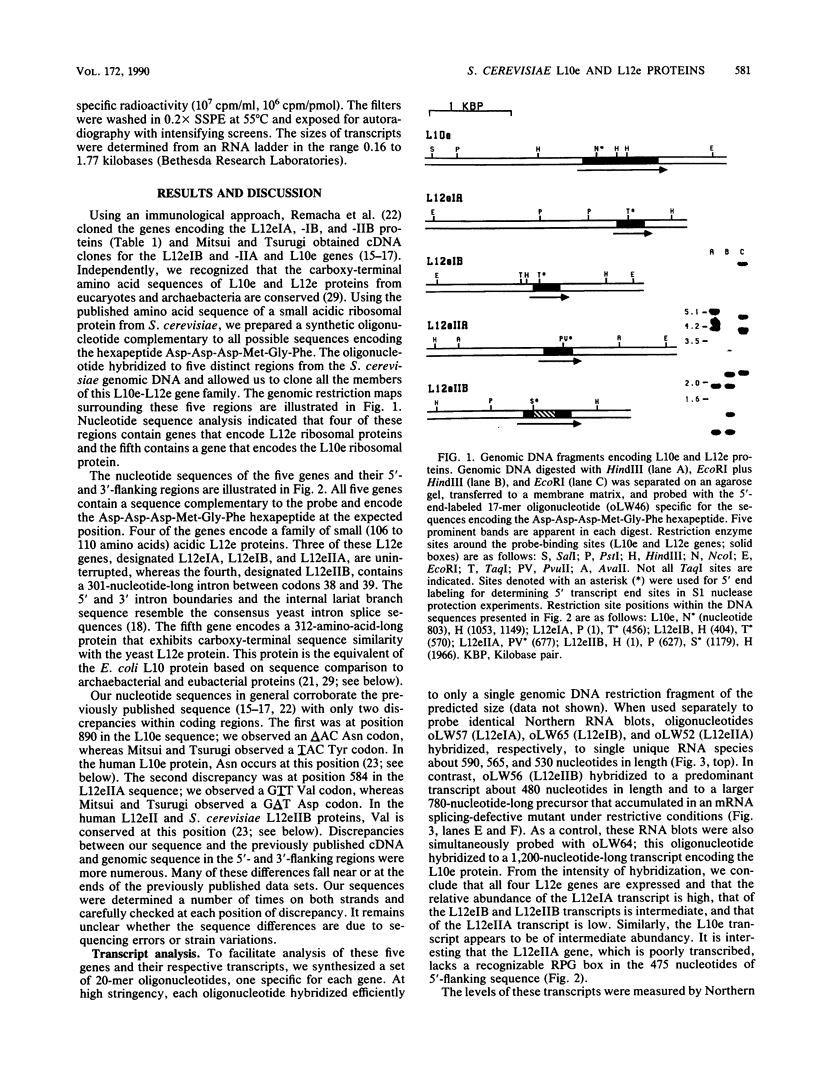
The budding yeast Saccharomyces cerevisiae contains a family of genes that encodes four different but related small acidic ribosomal proteins designated L12eIA, L12eIB, L12eIIA, and L12eIIB and a single larger protein designated L10e. These proteins are equivalent (e) to the L12 and L10 proteins of Escherichia coli that assemble as a 4:1 complex onto the large ribosomal subunit. The five yeast genes (or their cDNAs) have been cloned and sequenced (M. Remacha, M. T. Saenz-Robles, M. D. Vilella, and J. P. G. Ballesta, J. Biol. Chem. 263:9044-9101, 1988; K. Mitsui and K. Tsurugi, Nucleic Acids Res. 16:3573, 3574, and 3575, 1988; this work). Here, the transcripts of these genes were characterized and quantitated and the proteins they encode were compared and aligned. Four of the genes, L12eIA, -IB, -IIA, and L10e, are uninterrupted, whereas the L12eIIB gene contains a 301-nucleotide-long intron between codons 38 and 39. The transcripts derived from each of these genes were analyzed by Northern (RNA) hybridization, primer extension, and S1 nuclease protection. All five genes are expressed, albeit at different levels. The transcript levels are coordinate and exhibit growth rate-dependent regulation in rich (glucose) and poor (ethanol) media. The five yeast proteins each contain a highly conserved acidic carboxy terminus of about 20 residues in length. This domain of unknown function is also present in archaebacterial but absent from eubacterial L10e and L12e proteins. Comparisons of the factor-binding domains in the yeast and other eucaryotic and archaebacterial L12e proteins indicate that the original duplication to produce the type I and II genes was a very ancient event. The evolutionary relationships between the eucaryotic, archaebacterial, and eubacterial L10e and L12e genes (and proteins) are discussed.
Full text
PDF




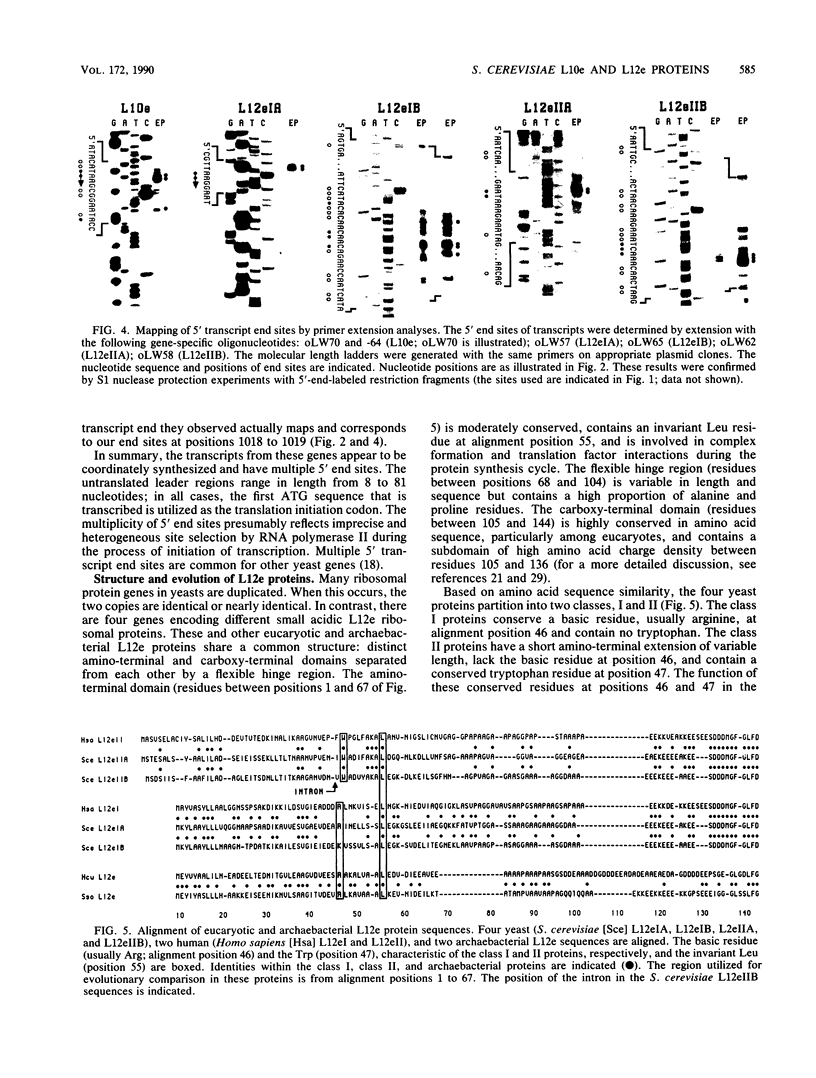
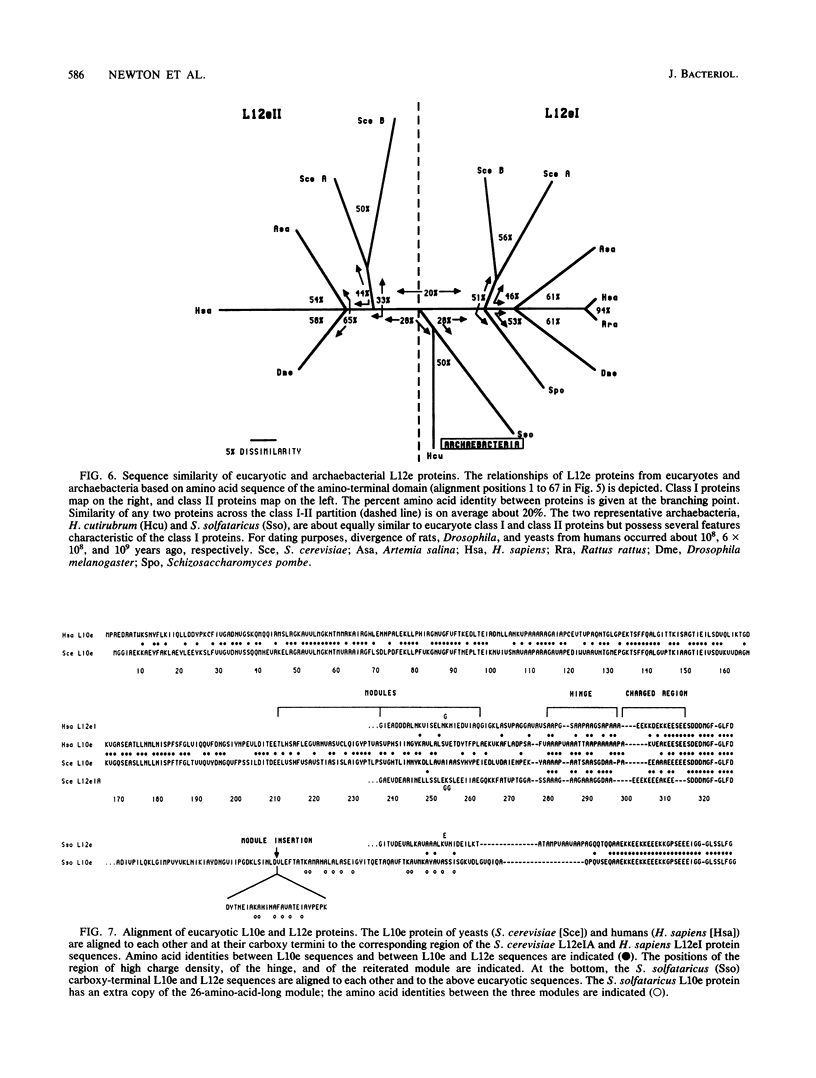


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amons R., Pluijms W., Möller W. The primary structure of ribosomal protein eL12/eL12-P from Artemia salina 80 S ribosomes. FEBS Lett. 1979 Aug 1;104(1):85–89. doi: 10.1016/0014-5793(79)81089-3. [DOI] [PubMed] [Google Scholar]
- Beltrame M., Bianchi M. E. Sequence of the cDNA for one acidic ribosomal protein of Schizosaccharomyces pombe. Nucleic Acids Res. 1987 Nov 11;15(21):9089–9089. doi: 10.1093/nar/15.21.9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Itoh T. Primary structure of an acidic ribosomal protein YPA1 from Saccharomyces cerevisiae. Isolation and characterization of peptides and the complete amino acid sequence. Biochim Biophys Acta. 1981 Nov 30;671(1):16–24. doi: 10.1016/0005-2795(81)90088-x. [DOI] [PubMed] [Google Scholar]
- Liljas A. Structural studies of ribosomes. Prog Biophys Mol Biol. 1982;40(3):161–228. doi: 10.1016/0079-6107(82)90013-x. [DOI] [PubMed] [Google Scholar]
- Lin A., Wittmann-Liebold B., McNally J., Wool I. G. The primary structure of the acidic phosphoprotein P2 from rat liver 60 S ribosomal subunits. Comparison with ribosomal 'A' proteins from other species. J Biol Chem. 1982 Aug 10;257(15):9189–9197. [PubMed] [Google Scholar]
- Miller A. M. The yeast MATa1 gene contains two introns. EMBO J. 1984 May;3(5):1061–1065. doi: 10.1002/j.1460-2075.1984.tb01927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tsurugi K. cDNA and deduced amino acid sequence of 38 kDa-type acidic ribosomal protein A0 from Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Apr 25;16(8):3573–3573. doi: 10.1093/nar/16.8.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tsurugi K. cDNA and deduced amino acid sequence of acidic ribosomal protein A1 from Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Apr 25;16(8):3574–3574. doi: 10.1093/nar/16.8.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tsurugi K. cDNA and deduced amino acid sequence of acidic ribosomal protein A2 from Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Apr 25;16(8):3575–3575. doi: 10.1093/nar/16.8.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Strycharz G. D., Nomura M., Lewis H., Dennis P. P. Nucleotide sequence of the ribosomal protein gene cluster adjacent to the gene for RNA polymerase subunit beta in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1697–1701. doi: 10.1073/pnas.76.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S., Zhang J. Y., Kay M. A., Jacobs-Lorena M. Structural analysis of the Drosophila rpA1 gene, a member of the eucaryotic 'A' type ribosomal protein family. Nucleic Acids Res. 1987 Feb 11;15(3):987–1003. doi: 10.1093/nar/15.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez C., Shimmin L. C., Newton C. H., Matheson A. T., Dennis P. P. Structure and evolution of the L11, L1, L10, and L12 equivalent ribosomal proteins in eubacteria, archaebacteria, and eucaryotes. Can J Microbiol. 1989 Jan;35(1):234–244. doi: 10.1139/m89-036. [DOI] [PubMed] [Google Scholar]
- Remacha M., Sáenz-Robles M. T., Vilella M. D., Ballesta J. P. Independent genes coding for three acidic proteins of the large ribosomal subunit from Saccharomyces cerevisiae. J Biol Chem. 1988 Jul 5;263(19):9094–9101. [PubMed] [Google Scholar]
- Rich B. E., Steitz J. A. Human acidic ribosomal phosphoproteins P0, P1, and P2: analysis of cDNA clones, in vitro synthesis, and assembly. Mol Cell Biol. 1987 Nov;7(11):4065–4074. doi: 10.1128/mcb.7.11.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M., Harris P. K., Woolford J. L., Jr, Teem J. L. The effect of temperature-sensitive RNA mutants on the transcription products from cloned ribosomal protein genes of yeast. Cell. 1981 Jun;24(3):679–686. doi: 10.1016/0092-8674(81)90094-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmin L. C., Dennis P. P. Characterization of the L11, L1, L10 and L12 equivalent ribosomal protein gene cluster of the halophilic archaebacterium Halobacterium cutirubrum. EMBO J. 1989 Apr;8(4):1225–1235. doi: 10.1002/j.1460-2075.1989.tb03496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmin L. C., Newton C. H., Ramirez C., Yee J., Downing W. L., Louie A., Matheson A. T., Dennis P. P. Organization of genes encoding the L11, L1, L10, and L12 equivalent ribosomal proteins in eubacteria, archaebacteria, and eucaryotes. Can J Microbiol. 1989 Jan;35(1):164–170. doi: 10.1139/m89-025. [DOI] [PubMed] [Google Scholar]
- Shimmin L. C., Ramirez C., Matheson A. T., Dennis P. P. Sequence alignment and evolutionary comparison of the L10 equivalent and L12 equivalent ribosomal proteins from archaebacteria, eubacteria, and eucaryotes. J Mol Evol. 1989 Nov;29(5):448–462. doi: 10.1007/BF02602915. [DOI] [PubMed] [Google Scholar]
- Stiles J. I. Use of integrative transformation of yeast in the cloning of mutant genes and large segments of contiguous chromosomal sequences. Methods Enzymol. 1983;101:290–300. doi: 10.1016/0076-6879(83)01022-8. [DOI] [PubMed] [Google Scholar]
- Strobel O., Köpke A. K., Kamp R. M., Böck A., Wittmann-Liebold B. Primary structure of the archaebacterial Methanococcus vannielii ribosomal protein L12. Amino acid sequence determination, oligonucleotide hybridization, and sequencing of the gene. J Biol Chem. 1988 May 15;263(14):6538–6546. [PubMed] [Google Scholar]
- Sánchez-Madrid F., Reyes R., Conde P., Ballesta J. P. Acidic ribosomal proteins from eukaryotic cells. Effect on ribosomal functions. Eur J Biochem. 1979 Aug 1;98(2):409–416. doi: 10.1111/j.1432-1033.1979.tb13200.x. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wigboldus J. D. cDNA and deduced amino acid sequence of Drosophila rp21C, another 'A'-type ribosomal protein. Nucleic Acids Res. 1987 Dec 10;15(23):10064–10064. doi: 10.1093/nar/15.23.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]