Abstract
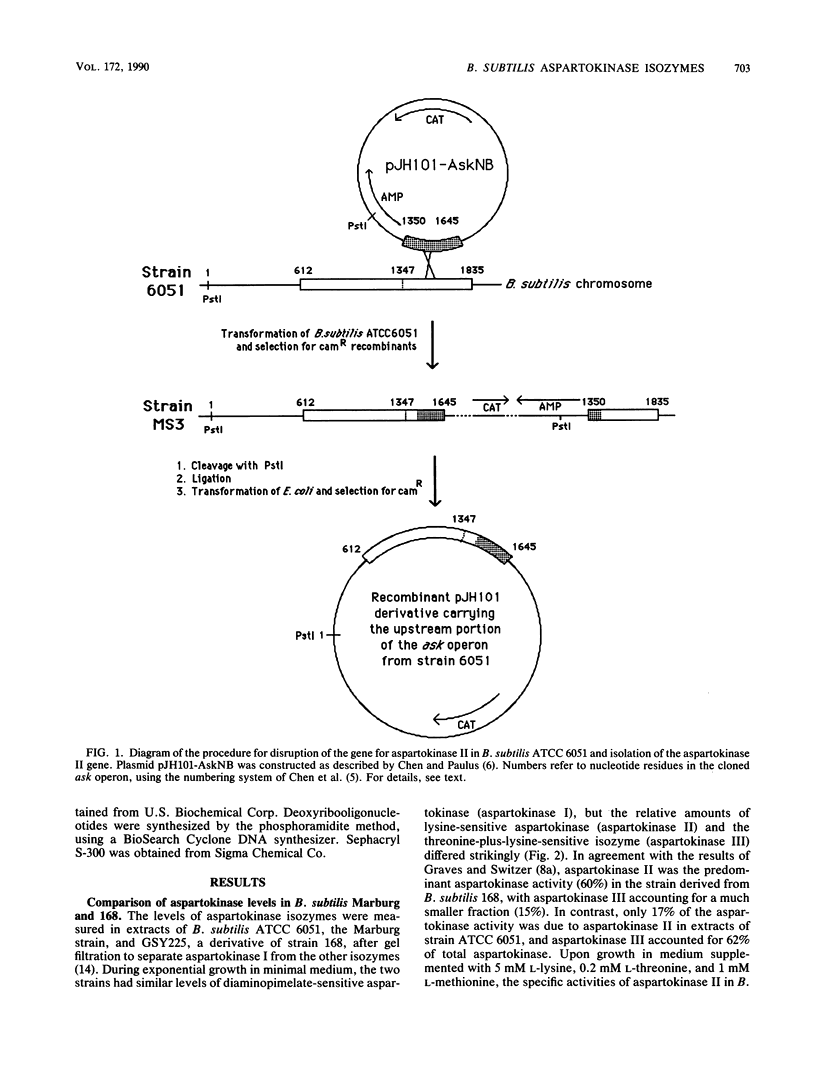
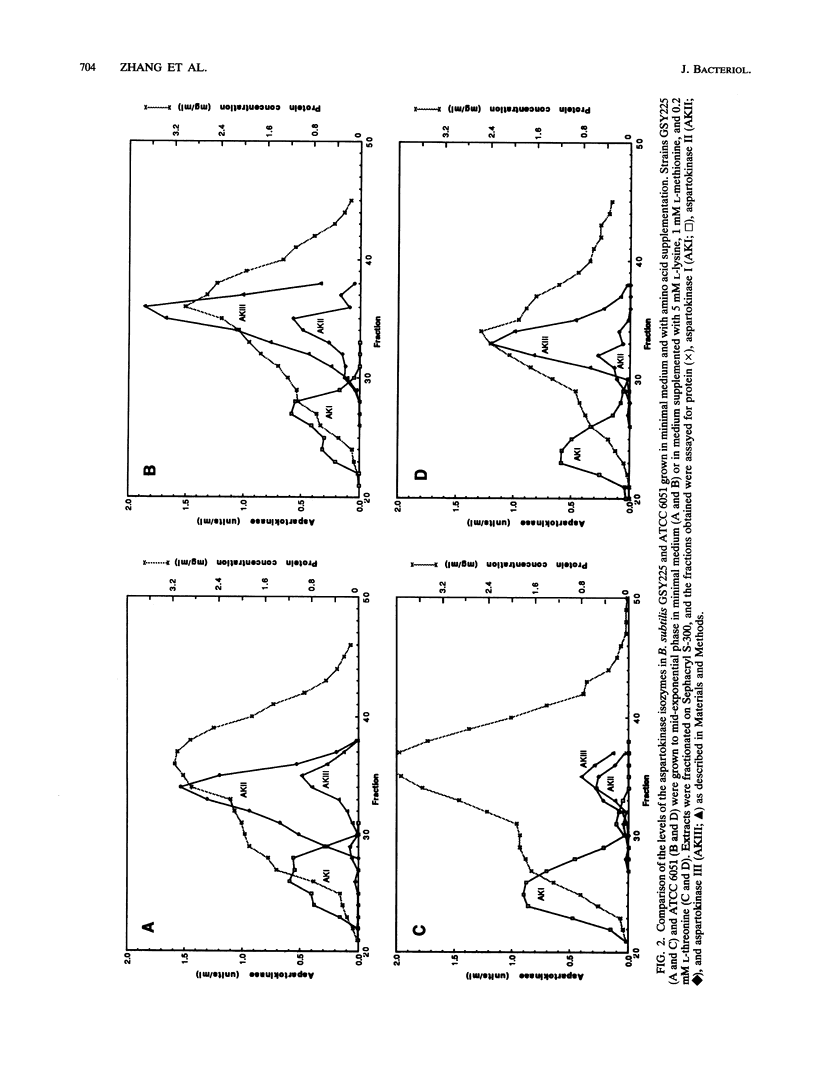
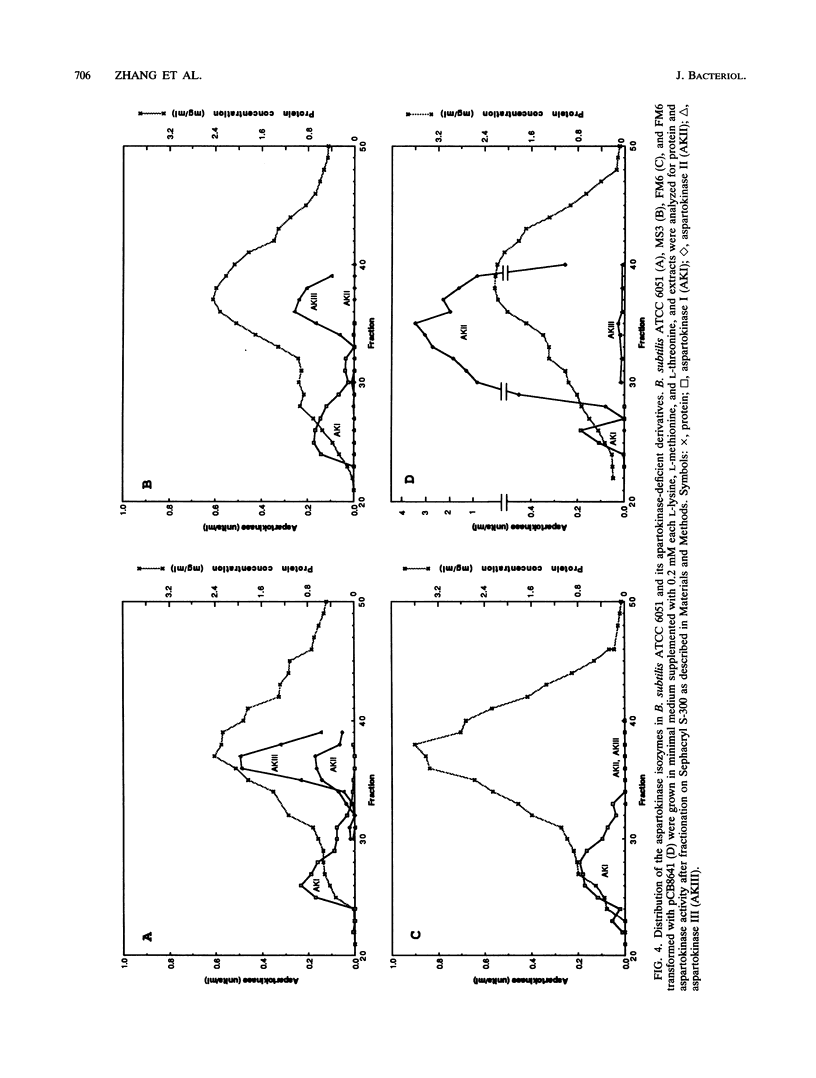
The levels of two aspartokinase isozymes, a lysine-sensitive enzyme and an aspartokinase that is inhibited synergistically by lysine plus threonine, differ strikingly in different strains of Bacillus subtilis. In derivatives of B. subtilis 168 growing in minimal medium, the predominant isozyme is the lysine-sensitive aspartokinase. In B. subtilis ATCC 6051, the Marburg strain, the level of the lysine-sensitive aspartokinase is much lower during growth in minimal medium, and the major aspartokinase activity is the lysine-plus-threonine-sensitive isozyme. Molecular cloning and nucleotide sequence determination of the genes for the lysine-sensitive isozymes from the two B. subtilis strains and their upstream control regions showed these genes to be identical. Evidence that the lysine-sensitive aspartokinase, referred to as aspartokinase II, is distinct from the threonine-plus-lysine-sensitive aspartokinase comes from the observation that disruption of the aspartokinase II gene by recombinational insertion had no effect on the latter. Mutants were obtained from the aspartokinase II-negative strain that also lacked the threonine-plus-lysine-sensitive aspartokinase, which will be referred to as aspartokinase III. Aspartokinase II could be selectively restored to these mutants by transformation with plasmids carrying the aspartokinase II gene. Study of the growth properties of the various mutant strains showed that the loss of either aspartokinase II or aspartokinase III had no effect on growth in minimal medium but that the loss of both enzymes interfered with growth unless the medium was supplemented with the three major end products of the aspartate pathway. It appears, therefore, that aspartokinase I alone cannot provide adequate supplies of precursors for the synthesis of lysine, threonine, and methionine by exponentially growing cells.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondaryk R. P., Paulus H. Cloning and structure of the gene for the subunits of aspartokinase II from Bacillus subtilis. J Biol Chem. 1985 Jan 10;260(1):585–591. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen N. Y., Hu F. M., Paulus H. Nucleotide sequence of the overlapping genes for the subunits of Bacillus subtilis aspartokinase II and their control regions. J Biol Chem. 1987 Jun 25;262(18):8787–8798. [PubMed] [Google Scholar]
- Chen N. Y., Paulus H. Mechanism of expression of the overlapping genes of Bacillus subtilis aspartokinase II. J Biol Chem. 1988 Jul 5;263(19):9526–9532. [PubMed] [Google Scholar]
- Chen N. Y., Zhang J. J., Paulus H. Chromosomal location of the Bacillus subtilis aspartokinase II gene and nucleotide sequence of the adjacent genes homologous to uvrC and trx of Escherichia coli. J Gen Microbiol. 1989 Nov;135(11):2931–2940. doi: 10.1099/00221287-135-11-2931. [DOI] [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D., Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves L. M., Switzer R. L. Aspartokinase III, a new isozyme in Bacillus subtilis 168. J Bacteriol. 1990 Jan;172(1):218–223. doi: 10.1128/jb.172.1.218-223.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldenwang W. G., Banner C. D., Ollington J. F., Losick R., Hoch J. A., O'Connor M. B., Sonenshein A. L. Mapping a cloned gene under sporulation control by inserttion of a drug resistance marker into the Bacillus subtilis chromosome. J Bacteriol. 1980 Apr;142(1):90–98. doi: 10.1128/jb.142.1.90-98.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton M. L., McCormick N. G., Behforouz N. C., Freese E. Regulation of two aspartokinases in Bacillus subtilis. J Bacteriol. 1971 Dec;108(3):1129–1134. doi: 10.1128/jb.108.3.1129-1134.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D., Paulus H. Properties and subunit structure of aspartokinase II from Bacillus subtilis VB217. J Biol Chem. 1977 Jul 10;252(13):4648–4651. [PubMed] [Google Scholar]
- Rosner A., Paulus H. Regulation of aspartokinase in Bacillus subtilis. The separation and properties of two isofunctional enzymes. J Biol Chem. 1971 May 10;246(9):2965–2971. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E. C., Steinberg W. The effect of gene position, gene dosage and a regulatory mutation on the temporal sequence of enzyme synthesis accompanying outgrowth of Bacillus subtilis spores. Mol Gen Genet. 1978 Jan 17;158(3):287–296. doi: 10.1007/BF00267200. [DOI] [PubMed] [Google Scholar]