Abstract
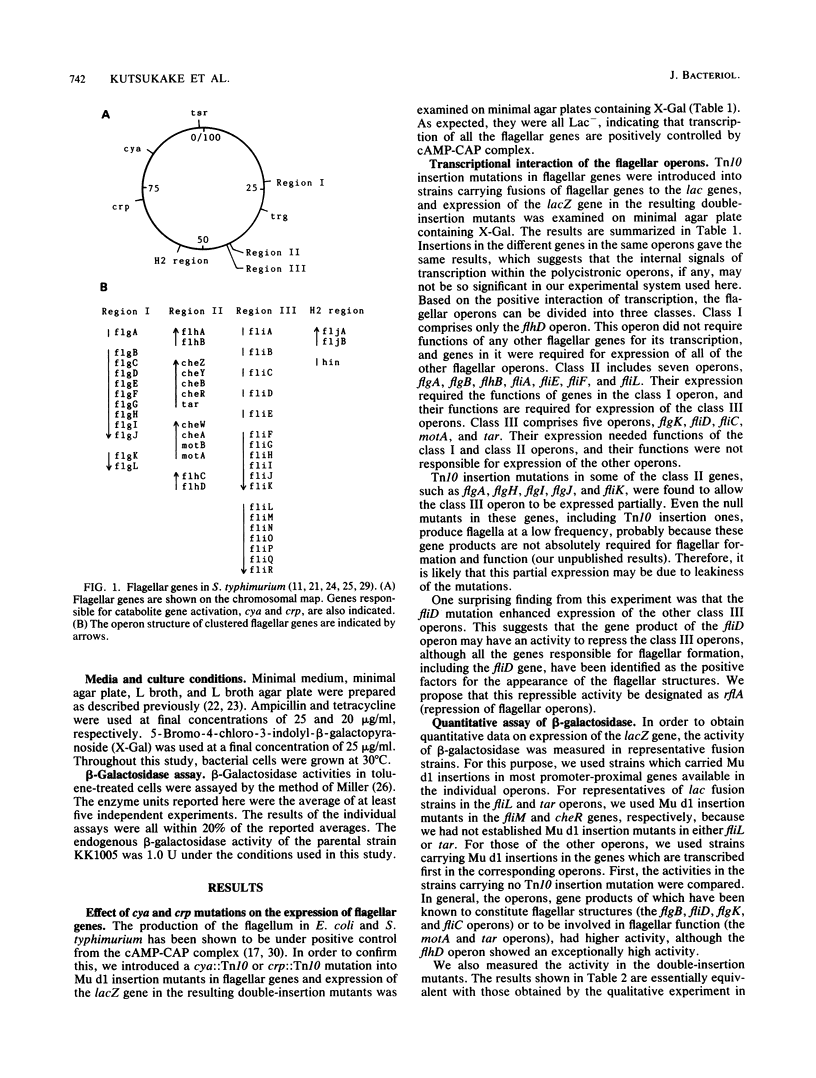
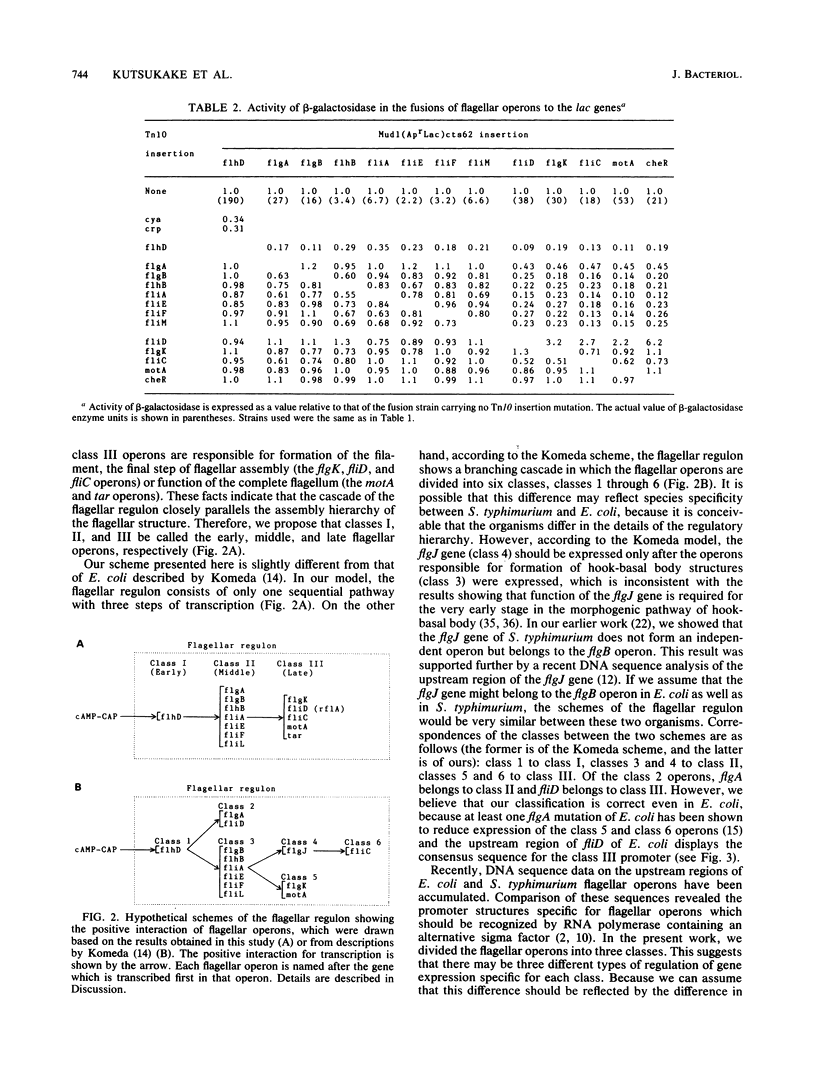
In Salmonella typhimurium, nearly 50 genes are involved in flagellar formation and function and constitute at least 13 different operons. In this study, we examined the transcriptional interaction among the flagellar operons by combined use of Mu d1(Apr Lac) cts62 and Tn10 insertion mutants in the flagellar genes. The results showed that the flagellar operons can be divided into three classes: class I contains only the flhD operon, which is controlled by the cAMP-CAP complex and is required for expression of all of the other flagellar operons; class II contains seven operons, flgA, flgB, flhB, fliA, fliE, fliF, and fliL, which are under control of class I and are required for the expression of class III; class III contains five operons, flgK, fliD fliC, motA, and tar. This ordered cascade of transcription closely parallels the assembly of the flagellar structure. In addition, we found that the fliD defect enhanced expression of the class III operons. This suggests that the fliD gene product may be responsible for repression of the class III operons in the mutants in the class II genes. These results are compared with the cascade model of the flagellar regulon of Escherichia coli proposed previously (Y. Komeda, J. Bacteriol. 170:1575-1581, 1982).
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnosti D. N., Chamberlin M. J. Secondary sigma factor controls transcription of flagellar and chemotaxis genes in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Feb;86(3):830–834. doi: 10.1073/pnas.86.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett D. H., Frantz B. B., Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5' consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988 Apr;170(4):1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger J., Park C., Harayama S., Hazelbauer G. L. Structure of the Trg protein: Homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champer R., Dingwall A., Shapiro L. Cascade regulation of Caulobacter flagellar and chemotaxis genes. J Mol Biol. 1987 Mar 5;194(1):71–80. doi: 10.1016/0022-2836(87)90716-9. [DOI] [PubMed] [Google Scholar]
- DeFranco A. L., Parkinson J. S., Koshland D. E., Jr Functional homology of chemotaxis genes in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1979 Jul;139(1):107–114. doi: 10.1128/jb.139.1.107-114.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G. E., Macnab R. M., Stader J., Matsumura P., Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984 Sep;159(3):991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa T., Sakai A., Tominaga A., Enomoto M. Isolation and characterization of Escherichia coli hag operator mutants whose hag48 expression has become repressible by a Salmonella H1 repressor. Mol Gen Genet. 1989 Mar;216(1):44–50. doi: 10.1007/BF00332229. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. DNA sequence analysis suggests that expression of flagellar and chemotaxis genes in Escherichia coli and Salmonella typhimurium is controlled by an alternative sigma factor. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6422–6424. doi: 10.1073/pnas.84.18.6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T., Komeda Y., Kutsukake K., Macnab R. M., Matsumura P., Parkinson J. S., Simon M. I., Yamaguchi S. New unified nomenclature for the flagellar genes of Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1988 Dec;52(4):533–535. doi: 10.1128/mr.52.4.533-535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Homma M., Macnab R. M. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jul;171(7):3890–3900. doi: 10.1128/jb.171.7.3890-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M., Homma M., Kutsukake K., Macnab R. M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jun;171(6):3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Fusions of flagellar operons to lactose genes on a mu lac bacteriophage. J Bacteriol. 1982 Apr;150(1):16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y., Kutsukake K., Iino T. Definition of additional flagellar genes in Escherichia coli K12. Genetics. 1980 Feb;94(2):277–290. doi: 10.1093/genetics/94.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y., Suzuki H., Ishidsu J. I., Iino T. The role of cAMP in flagellation of Salmonella typhimurium. Mol Gen Genet. 1976 Dec 31;142(4):289–298. doi: 10.1007/BF00271253. [DOI] [PubMed] [Google Scholar]
- Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986 Dec;168(3):1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Sequence of the flaA (cheC) locus of Escherichia coli and discovery of a new gene. J Bacteriol. 1986 Jun;166(3):1007–1012. doi: 10.1128/jb.166.3.1007-1012.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K., Iino T., Komeda Y., Yamaguchi S. Functional homology of fla genes between Salmonella typhimurium and Escherichia coli. Mol Gen Genet. 1980 Apr;178(1):59–67. doi: 10.1007/BF00267213. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Iino T. Refined genetic analysis of the region II che mutants in Salmonella typhimurium. Mol Gen Genet. 1985;199(3):406–409. doi: 10.1007/BF00330750. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Ohya Y., Yamaguchi S., Iino T. Operon structure of flagellar genes in Salmonella typhimurium. Mol Gen Genet. 1988 Sep;214(1):11–15. doi: 10.1007/BF00340172. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Suzuki T., Yamaguchi S., Iino T. Role of gene flaFV on flagellar hook formation in Salmonella typhimurium. J Bacteriol. 1979 Oct;140(1):267–275. doi: 10.1128/jb.140.1.267-275.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A., Ohta N., Ramakrishnan G., Mullin D., Raymond G. Genetic switching in the flagellar gene hierarchy of Caulobacter requires negative as well as positive regulation of transcription. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6651–6655. doi: 10.1073/pnas.86.17.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. I. Bacterial flagella. Annu Rev Microbiol. 1977;31:397–419. doi: 10.1146/annurev.mi.31.100177.002145. [DOI] [PubMed] [Google Scholar]
- Silverman M., Simon M. Characterization of Escherichia coli flagellar mutants that are insensitive to catabolite repression. J Bacteriol. 1974 Dec;120(3):1196–1203. doi: 10.1128/jb.120.3.1196-1203.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocum M. K., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J Bacteriol. 1983 Aug;155(2):565–577. doi: 10.1128/jb.155.2.565-577.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Iino T. Absence of messenger ribonucleic acid specific for flagellin in non-flagellate mutants of Salmonella. J Mol Biol. 1975 Jul 15;95(4):549–556. doi: 10.1016/0022-2836(75)90316-2. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Iino T., Horiguchi T., Yamaguchi S. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J Bacteriol. 1978 Feb;133(2):904–915. doi: 10.1128/jb.133.2.904-915.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Komeda Y. Incomplete flagellar structures in Escherichia coli mutants. J Bacteriol. 1981 Feb;145(2):1036–1041. doi: 10.1128/jb.145.2.1036-1041.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely E., Simon M. DNA sequence adjacent to flagellar genes and evolution of flagellar-phase variation. J Bacteriol. 1983 Jul;155(1):74–81. doi: 10.1128/jb.155.1.74-81.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Dingwall A., Shapiro L. Negative transcriptional regulation in the Caulobacter flagellar hierarchy. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6656–6660. doi: 10.1073/pnas.86.17.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Fujita H., Taira T., Kutsukake K., Homma M., Iino T. Genetic analysis of three additional fla genes in Salmonella typhimurium. J Gen Microbiol. 1984 Dec;130(12):3339–3342. doi: 10.1099/00221287-130-12-3339. [DOI] [PubMed] [Google Scholar]
- Zieg J., Simon M. Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]



