Abstract
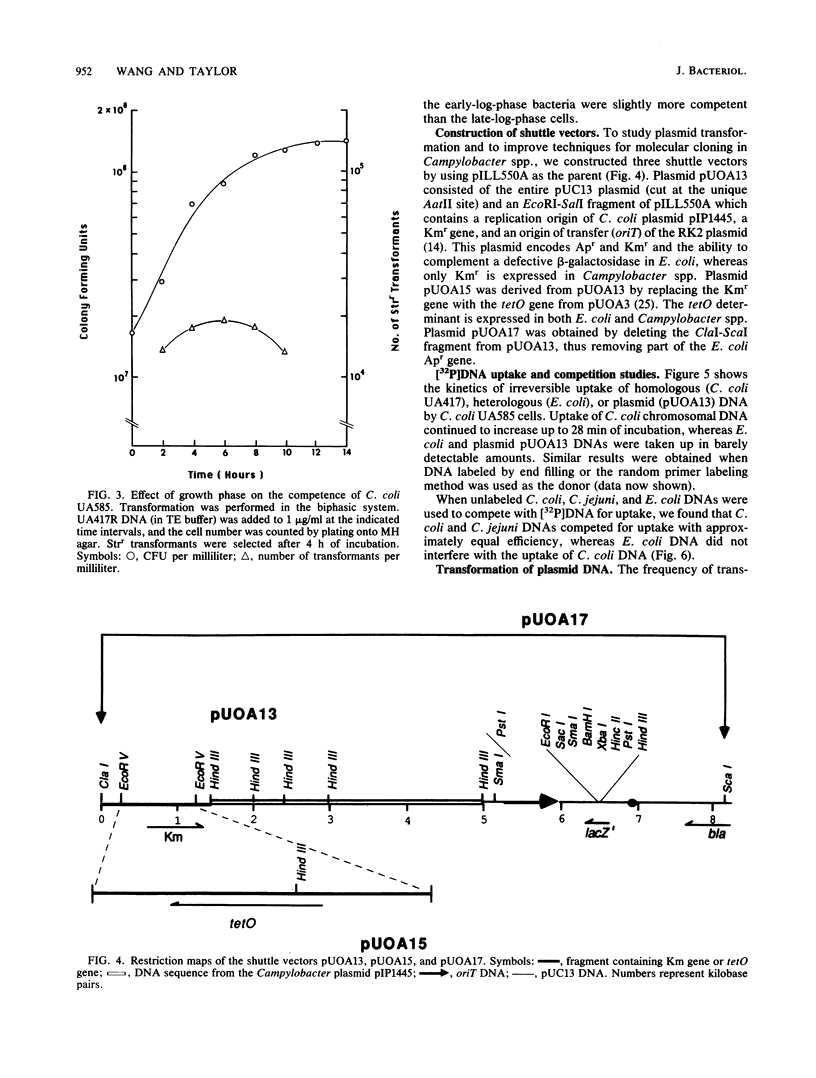
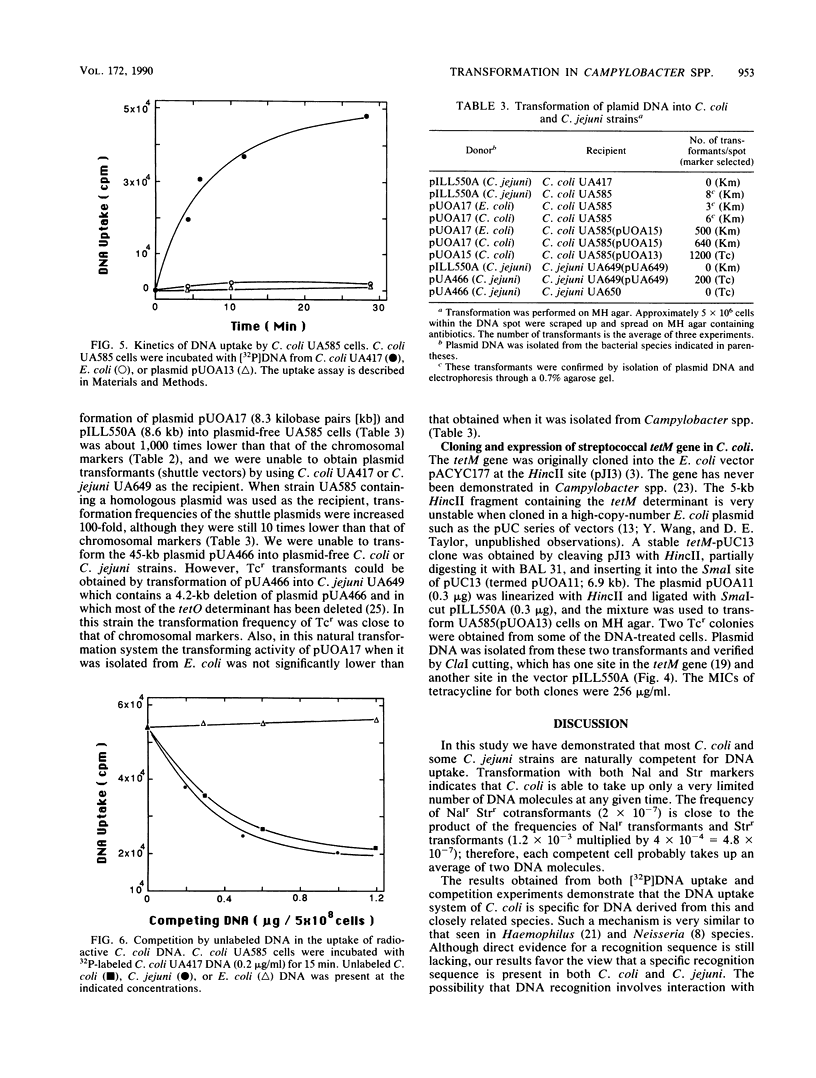
Growing cells of Campylobacter coli and C. jejuni were naturally transformed by naked DNA without the requirement for any special treatment. Transformation frequencies for homologous chromosomal DNA were approximately 10(-3) transformants per recipient cell in C. coli and 10(-4) in C. jejuni. Maximum competence was found in the early log phase of growth. Campylobacters preferentially took up their own DNA in comparison with Escherichia coli chromosomal DNA, which was taken up very poorly. Three new Campylobacter spp.-to-E. coli shuttle plasmids, which contained additional cloning sites and selectable markers, were constructed from the shuttle vector pILL550A. These plasmid DNAs were taken up by campylobacters much less efficiently than was homologous chromosomal DNA, and transformation into plasmid-free cells was very rare. However, with the use of recipients containing a homologous plasmid, approximately 10(-4) transformants per cell were obtained. The tetM determinant, originally obtained from Streptococcus spp. and not heretofore reported in Campylobacter spp., was isolated from an E. coli plasmid and was introduced, selecting for tetracycline resistance, by natural transformation into C. coli.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belland R. J., Trust T. J. Deoxyribonucleic acid sequence relatedness between thermophilic members of the genus Campylobacter. J Gen Microbiol. 1982 Nov;128(11):2515–2522. doi: 10.1099/00221287-128-11-2515. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett V., Inamine J., Rajagopalan S. Heterogeneity of tetracycline resistance determinants in Streptococcus. J Bacteriol. 1982 Mar;149(3):995–1004. doi: 10.1128/jb.149.3.995-1004.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Chung C. T., Niemela S. L., Miller R. H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner D. B., Deich R. A., Sisco K. L., Smith H. O. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene. 1980 Nov;11(3-4):311–318. doi: 10.1016/0378-1119(80)90071-2. [DOI] [PubMed] [Google Scholar]
- Dougherty T. J., Asmus A., Tomasz A. Specificity of DNA uptake in genetic transformation of gonococci. Biochem Biophys Res Commun. 1979 Jan 15;86(1):97–104. doi: 10.1016/0006-291x(79)90386-3. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Goodgal S. H. DNA uptake in Haemophilus transformation. Annu Rev Genet. 1982;16:169–192. doi: 10.1146/annurev.ge.16.120182.001125. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Scocca J. J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Venema G., Daly C., Fitzgerald G. F. Cloning and characterization of the tetracycline resistance determinant of and several promoters from within the conjugative transposon Tn919. Appl Environ Microbiol. 1988 May;54(5):1230–1236. doi: 10.1128/aem.54.5.1230-1236.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labigne-Roussel A., Harel J., Tompkins L. Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J Bacteriol. 1987 Nov;169(11):5320–5323. doi: 10.1128/jb.169.11.5320-5323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Wood P. H., Jones K. R. Genetic transformation of Streptococcus sanguis (Challis) with cryptic plasmids from Streptococcus ferus. Infect Immun. 1980 Jun;28(3):692–699. doi: 10.1128/iai.28.3.692-699.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P., Trieu-Cuot P., Courvalin P. Nucleotide sequence of the tetM tetracycline resistance determinant of the streptococcal conjugative shuttle transposon Tn1545. Nucleic Acids Res. 1986 Sep 11;14(17):7047–7058. doi: 10.1093/nar/14.17.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. F., Dower W. J., Tompkins L. S. High-voltage electroporation of bacteria: genetic transformation of Campylobacter jejuni with plasmid DNA. Proc Natl Acad Sci U S A. 1988 Feb;85(3):856–860. doi: 10.1073/pnas.85.3.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scocca J. J., Poland R. L., Zoon K. C. Specificity in deoxyribonucleic acid uptake by transformable Haemophilus influenzae. J Bacteriol. 1974 May;118(2):369–373. doi: 10.1128/jb.118.2.369-373.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Danner D. B., Deich R. A. Genetic transformation. Annu Rev Biochem. 1981;50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- Stewart G. J., Carlson C. A. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–235. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- Taylor D. E., Chang N., Garner R. S., Sherburne R., Mueller L. Incidence of antibiotic resistance and characterization of plasmids in Campylobacter jejuni strains isolated from clinical sources in Alberta, Canada. Can J Microbiol. 1986 Jan;32(1):28–32. doi: 10.1139/m86-006. [DOI] [PubMed] [Google Scholar]
- Taylor D. E., Hiratsuka K., Ray H., Manavathu E. K. Characterization and expression of a cloned tetracycline resistance determinant from Campylobacter jejuni plasmid pUA466. J Bacteriol. 1987 Jul;169(7):2984–2989. doi: 10.1128/jb.169.7.2984-2989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. E., Ng L. K., Lior H. Susceptibility of Campylobacter species to nalidixic acid, enoxacin, and other DNA gyrase inhibitors. Antimicrob Agents Chemother. 1985 Nov;28(5):708–710. doi: 10.1128/aac.28.5.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Gerbaud G., Lambert T., Courvalin P. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 1985 Dec 16;4(13A):3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]



