Abstract
One of the most provocative recent discoveries in immunology was the description of a genetic linkage in the major histocompatibility complex (MHC) between structurally unrelated genes whose products are involved in processing and presentation of antigens for recognition by T lymphocytes. Genes encoding MHC class I molecules, which bind and present at the cell surface proteolytic fragments of cytosolic proteins, are linked to nonhomologous genes whose products are involved in the production and subsequent transfer of such fragments into the endoplasmic reticulum. In mammals, the class I presentation and processing genes are found in different regions of the MHC. To examine the evolutionary origins of this genetic association, linkage studies were carried out with Xenopus, an amphibian last sharing an ancestor with mammals over 350 million years ago. In contrast to mammals, the single copy Xenopus class I gene is located between the class II and III regions, speculated to be in close linkage with the processing and transport genes. In addition to suggesting a primordial organization of genes involved in class I antigen presentation, these linkage studies further provide insight into the origins of the MHC class III region and the phenomenon of class I gene instability in the mammalian MHC.
Keywords: evolution, linkage groups, antigen presentation
The major histocompatibility complex (MHC), whose major function is to initiate adaptive immune responses in vertebrates, is a large genetic region encompassing more than 4000 kb in humans (1). As defined in mammals, the MHC is comprised of three regions, classes I, II, and III (see Fig. 2). The class II region contains loci specifying the canonical peptide-binding molecules expressed by antigen-presenting cells and also includes genes encoding proteasome components [low molecular weight proteins (lmp) 2 and 7] and endoplasmic reticulum transporters (transporters associated with antigen processing) required for the production and transport of antigenic peptides that bind to class I molecules. The class III region or “region of unrelated loci” contains many genes, some of which are involved in immunity but apparently not directly in antigen presentation (2, 3). The class I region in different mammalian species contains a wildly varying number of class I genes, one to three of which are polymorphic and are expressed ubiquitously (class Ia) and others that are monomorphic that are either pseudogenes or may serve restricted functions (class Ib; ref. 4). The class II and III regions are relatively conserved in evolution; orthologous genes are easily detected among mammalian taxa; in contrast, it is difficult or impossible to identify functional orthologous class I genes even among certain primate groups (1, 5–7). Thus, two major questions have arisen: (i) why are class I antigen processing genes in the class II region? and (ii) why is the class I region unstable relative to classes II and III?
Figure 2.

Gene order in the MHC of most mammals and the proposed order of the primordial MHC. The hypothesis is that class I originally was (and is, in some taxa) closely linked to the genes whose products are involved in the processing and transport of peptides to be presented by class I proteins. This original grouping of genes may have allowed for maintenance of certain allelic lineages of class I and class I-processing genes (1, 22). Subsequent translocation of class I distal to the complex made class I more prone to duplicate and diverge, the situation found in many mammalian species.
The Xenopus MHC is remarkably similar to its mouse and human counterparts (8). Classes Ia (9), IIβ (10), and class IIα (ref. 11; Lin, Y., and M.F.F., unpublished work), the complement components factor B (Bf; ref. 12) and C4 (13, 14), heat shock protein 70 (HSP70; ref. 15), the proteasome element lmp7 (16), and RING3 (17) genes all map to the Xenopus MHC, demonstrating that the the same class I, II, and III components were genetically linked when amphibians and mammals diverged from an ancestor 300–350 million years ago (Fig. 1b). Furthermore, previously we detected an intra-MHC recombinant in family studies that demonstrated that the Xenopus class II/lmp7 genes could be separated from the class Ia/Bf/HSP70 genes (16), which, at first glance, suggested that even the gene order of the Xenopus MHC is similar to that of mammals (recombinant noted in the top line of Fig. 1b). One significant difference between the Xenopus MHC and those of mammals is the extraordinary stability of the MHC-linked class I gene; in all clawed frog species analyzed to date, some separated for up to 100 million years, only one class I gene, of the classical type, is present in each haplotype (9). A large number of class Ib genes is indeed present in Xenopus, but these are not linked to the MHC proper (19).
Figure 1.
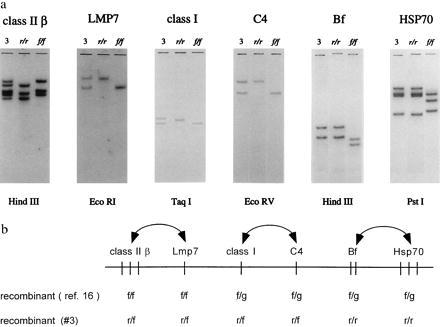
Southern blot analysis of intra-MHC recombinants suggests that, in contrast to mammals, the Xenopus MHC class Ia gene is associated with the class II region. (a) Southern blotting analysis of class IIβ (10), lmp7 (16), class Ia (9), C4 (14), Bf (12), and HSP70 (15) genes using DNA from 17 members of an r/f × r/f cross. Probes detected an intra-MHC recombinant (animal #3). The r/r and f/f lanes display the hybridization patterns of the f and r MHC haplotypes (DNA obtained from MHC homozygotes). (b) Order of Xenopus MHC genes based on recombinants (barring double crossovers) detected in pedigree analyses. In previous tying of members of an f/f × f/g family, recombinant offspring #15 displayed cosegregation of class II and lmp7 on the one hand and class Ia, C4, Bf, C4, and HSP70 on the other (16). The recombinant haplotypes noted below are derived from the animal in Fig. 1a. Vertical bars indicate genes. Doubleheaded arrows show that the order of the indicated loci is unknown and may be reversed. Note that the number of HSP70 genes may be two or three (15).
In this study, we carried out further mapping studies that strongly suggest that, in contrast to the situation in mammals, the class Ia gene is located between the class II and III regions. Such data imply that the genes involved in the processing and transport of peptides were originally closely linked to the class I “presenting” genes. In addition, our work further supports the premise of an ancient evolutionary association of the class III region and the MHC.
MATERIALS AND METHODS
Animals.
Xenopus laevis bearing the r, f, or g MHC haplotypes (8) were used. Two crosses were performed: r/f × r/f and f/f × f/g (16). Offspring in these families were analyzed for segregation of the MHC-linked class IIβ, lmp7, class Ia, C4, Bf, and HSP70 genes as described (16).
Southern Blotting.
One or two micrograms of genomic DNA, isolated from erythrocytes of individual adult offspring in the two families, was digested with the indicated restriction enzymes noted in Fig. 1, and Southern hybridizations were carried out as described (12, 16).
RESULTS AND DISCUSSION
Gene Order in the Xenopus MHC.
Using gene probes for class IIβ, lmp7, class Ia, C4, Bf, and HSP70, perfect cosegregation was detected among offspring of an r/f × r/f family except in one case. In this intra-MHC recombinant (#3), the class II/lmp7/class Ia/C4 genes cosegregated away from the Bf/HSP70 loci (Fig. 1a). Barring the unlikely event of a double crossover, this recombinant, taken together with the data from a previous family study (Fig. 1b; ref. 16), defined three regions of the Xenopus MHC: class IIβ/lmp7, class Ia/C4, and Bf/HSP70. Although the exact gene order is yet to be resolved, one intriguing possibility (that we favor) is that the lmp7 and class Ia genes are adjacent to each other as are the C4 and Bf genes, providing two examples of a close linkage between functionally related but structurally unrelated genes. This result is gratifying because the class I processing genes are found in the class II region in all species so far examined (1). Furthermore, class I and II genes clearly have been derived from a common ancestor (20, 21) at some point in evolution, so a close linkage between them is expected.
Class I Gene Plasticity and Origin of the Class III Region.
A more intimate association of class I with the class II/III regions (and by inference a close association of class I with the processing/transport genes; Fig. 2) also may explain the remarkably deep and stable MHC-linked class Ia lineage in Xenopus; we propose that the translocation of class I out of this relatively stable, “protected” genetic region to the distal end of the MHC, later in vertebrate evolution, bestowed on class I genes the capacity to duplicate and diverge, hence giving rise to the celebrated (and perhaps advantageous) class I plasticity found in mammals (4, 22). We predict that living representatives of other nonmammalian vertebrate classes also may display this “primordial” linkage pattern; data to date suggest that, similar to Xenopus but in contrast to mammals, there are deep lineages of teleost class I genes (refs. 23, 24, and Parham, P., personal communication). In addition, preliminary data suggest that zebrafish class I genes are closely linked to proteasome (lmp) genes (Vincek and Graser, personal communication). In one other case, Xenopus genes appear to be in a primordial organization; the globin gene cluster contains both the homologous α and β genes whereas in higher vertebrates separate α and β clusters exist (25).
Another implication of our data is the possible physiological significance of the linkage of the class III genes to MHC. The class III region has maintained its association with class I and class II despite gross genetic reorganizations that occurred over evolutionary time. Thus, it is very unlikely that the class III region of mammals was inserted between the class I and class II regions in a “genetic accident” (26). Indeed, from mapping studies in humans and mice, Kasahara et al. (27) recently proposed that the primordial MHC was comprised of (at least) ancestral class I processing genes and several class III-like genes.
Coselection of Allelic Lineages?
Our work suggests that, because of the close linkage of class I to the processing genes, certain “sets” of proteasome/transporter/class I lineages could have been selected and perpetuated, i.e., a close linkage between the class I and processing genes may have facilitated coselection of alleles derived from the different loci (1, 22). This hypothesis can be tested in Xenopus, in which certain divergent lmp7 alleles are linked to equally diverse class I alleles (ref. 16; M.F.F., unpublished work). Such coselection may now be rare in mammals (e.g., the rat; ref. 28) because of their vast class I gene plasticity. In the chicken MHC, a peptide transporter gene is closely linked to (sandwiched between) the two expressed class Ia genes (29). Thus, despite the major reorganization of the bird MHC (e.g., an apparent complete loss of the class III region), the primordial association between class II and I regions seems to have been preserved in this taxon. The linkage of functionally related yet structurally unrelated genes is rare in eukaryotes; ongoing phylogenetic studies of this particular system may not only uncover the origins of the MHC but may also provide insight into general mechanisms of genome evolution.
Acknowledgments
We thank Louis Du Pasquier and Masanori Kasahara for discussion of the data and reading of the manuscript. M.N. is supported by the Ministry of Education, Science, and Culture of Japan, and M.F.F. is supported by the National Institutes of Health Grant AI27877.
ABBREVIATIONS
- MHC
major histocompatibility complex
- lmp
low molecular weight protein (inducible proteasome component)
- Bf
factor B
- HSP70
heat shock protein 70
References
- 1.Trowsdale J. Immunogenetics. 1995;7:1–17. doi: 10.1007/BF00188427. [DOI] [PubMed] [Google Scholar]
- 2.Klein J. Natural History of the MHC. New York: Wiley; 1986. [Google Scholar]
- 3.Campbell R D, Trowsdale J. Immunol Today. 1993;14:349–352. doi: 10.1016/0167-5699(93)90234-C. [DOI] [PubMed] [Google Scholar]
- 4.Kasahara M, Kandil E, Salter-Cid L, Flajnik M F. Res Immunol. 1996;147:278–285. doi: 10.1016/0923-2494(96)89640-4. [DOI] [PubMed] [Google Scholar]
- 5.Klein J, Figueroa F. Immunol Today. 1986;7:41–44. doi: 10.1016/0167-5699(86)90123-4. [DOI] [PubMed] [Google Scholar]
- 6.Parham P. Semin Immunol. 1994;6:373–382. doi: 10.1006/smim.1994.1047. [DOI] [PubMed] [Google Scholar]
- 7.Lawlor D A, Warren E, Ward F E, Parham P. Immunol Rev. 1990;113:148–185. doi: 10.1111/j.1600-065x.1990.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 8.Flajnik M F, Du Pasquier L. Immunol Rev. 1990;113:47–63. doi: 10.1111/j.1600-065x.1990.tb00036.x. [DOI] [PubMed] [Google Scholar]
- 9.Shum B P, Avila D, Du Pasquier L, Kasahara M, Flajnik M F. J Immunol. 1993;151:5376–5386. [PubMed] [Google Scholar]
- 10.Sato K, Flajnik M F, Du Pasquier L, Katagiri M, Kasahara M. J Immunol. 1993;150:2831–2843. [PubMed] [Google Scholar]
- 11.Kaufman J F, Flajnik M F, Du Pasquier L. J Immunol. 1985;134:3258–3264. [PubMed] [Google Scholar]
- 12.Kato Y, Salter-Cid L, Flajnik M F, Namikawa C, Sasaki M, Nonaka M. J Immunol. 1994;153:4546–4554. [PubMed] [Google Scholar]
- 13.Nakamura T, Sekizawa A, Fujii T, Katagiri C. Immunogenetics. 1986;23:181–186. doi: 10.1007/BF00373819. [DOI] [PubMed] [Google Scholar]
- 14.Mo R, Kato M, Nonaka M, Nakayama K, Takahashi M. Immunogenetics. 1996;43:360–369. doi: 10.1007/BF02199804. [DOI] [PubMed] [Google Scholar]
- 15.Salter-Cid L, Kasahara M, Flajnik M F. Immunogenetics. 1994;39:1–7. doi: 10.1007/BF00171790. [DOI] [PubMed] [Google Scholar]
- 16.Namikawa C, Salter-Cid L, Flajnik M F, Kato Y, Nonaka M, Sasaki M. J Immunol. 1995;155:1964–1971. [PubMed] [Google Scholar]
- 17.Salter-Cid L, Du Pasquier L, Flajnik M F. Immunogenetics. 1996;44:397–399. [PubMed] [Google Scholar]
- 18.Flajnik M F, Kaufman J F, Riegert P, Du Pasquier L. Immunogenetics. 1984;20:433–442. doi: 10.1007/BF00345617. [DOI] [PubMed] [Google Scholar]
- 19.Flajnik M F, Kasahara M, Shum B P, Salter-Cid L, Taylor E, Du Pasquier L. EMBO J. 1993;12:4385–4396. doi: 10.1002/j.1460-2075.1993.tb06123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman J F. Verh Dtsch Zool Ges. 1989;81:131–144. [Google Scholar]
- 21.Hughes A L, Nei M. Immunogenetics. 1993;37:337–346. doi: 10.1007/BF00216798. [DOI] [PubMed] [Google Scholar]
- 22.Kasahara M, Flajnik M F, Ishibashi T, Natori T. Transplant Immunol. 1995;3:1–20. doi: 10.1016/0966-3274(95)80001-8. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi H, Figueroa F, O’Huigin C, Klein J. Immunogenetics. 1995;42:77–84. doi: 10.1007/BF00178581. [DOI] [PubMed] [Google Scholar]
- 24.Van Erp S H M, Egberts E, Stet R J M. Eur J Immunogenetics. 1996;23:371–381. doi: 10.1111/j.1744-313x.1996.tb00010.x. [DOI] [PubMed] [Google Scholar]
- 25.Jeffreys A J, Wilson V, Wood D, Simons J P. Cell. 1980;21:555–564. doi: 10.1016/0092-8674(80)90493-6. [DOI] [PubMed] [Google Scholar]
- 26.Klein J, Figueroa F. Crit Rev Immunol. 1986;6:295–386. [PubMed] [Google Scholar]
- 27.Kasahara M, Hayashi M, Tanaka K, Inoko H, Sugaya K, Ikemura T, Ishibashi T. Proc Natl Acad Sci. 1996;93:9096–9101. doi: 10.1073/pnas.93.17.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livingstone A M, Powis S J, Günther C, Cramer D V, Howard J C, Butcher G W. Immunogenetics. 1991;34:157–163. doi: 10.1007/BF00205818. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman J, Völk H, Wallny H-J. Immunol Rev. 1995;143:63–88. doi: 10.1111/j.1600-065x.1995.tb00670.x. [DOI] [PubMed] [Google Scholar]


