Abstract
Previous genetic analyses of the Caulobacter crescentus chromosome have resulted in the construction of a linear genetic map. To establish the circularity of the C. crescentus chromosome, restriction fragments generated by digestion with AseI and SpeI were analyzed by pulsed-field gel electrophoresis and Southern hybridization. The size of each fragment was calculated and used to demonstrate that C. crescentus has a genome size of approximately 4,000 kilobases. In addition, both enzymes gave rise to large DNA fragments which contained genes from both ends of the genetic map. Thus, there is physical linkage between the genes at the ends of the genetic map and the chromosome is circular. Since this region of the chromosome appears to contain the replication terminus, we propose that recombination occurs at a high frequency in the vicinity of the terminus. This high frequency of recombination would prevent genetic linkage from being observed between genes on opposite sides of the terminus. Additional experiments using insertions which introduced new AseI and DraI restriction sites into the genome allowed us to calculate the physical distance between genes located in the vicinity of the replication terminus.
Full text
PDF
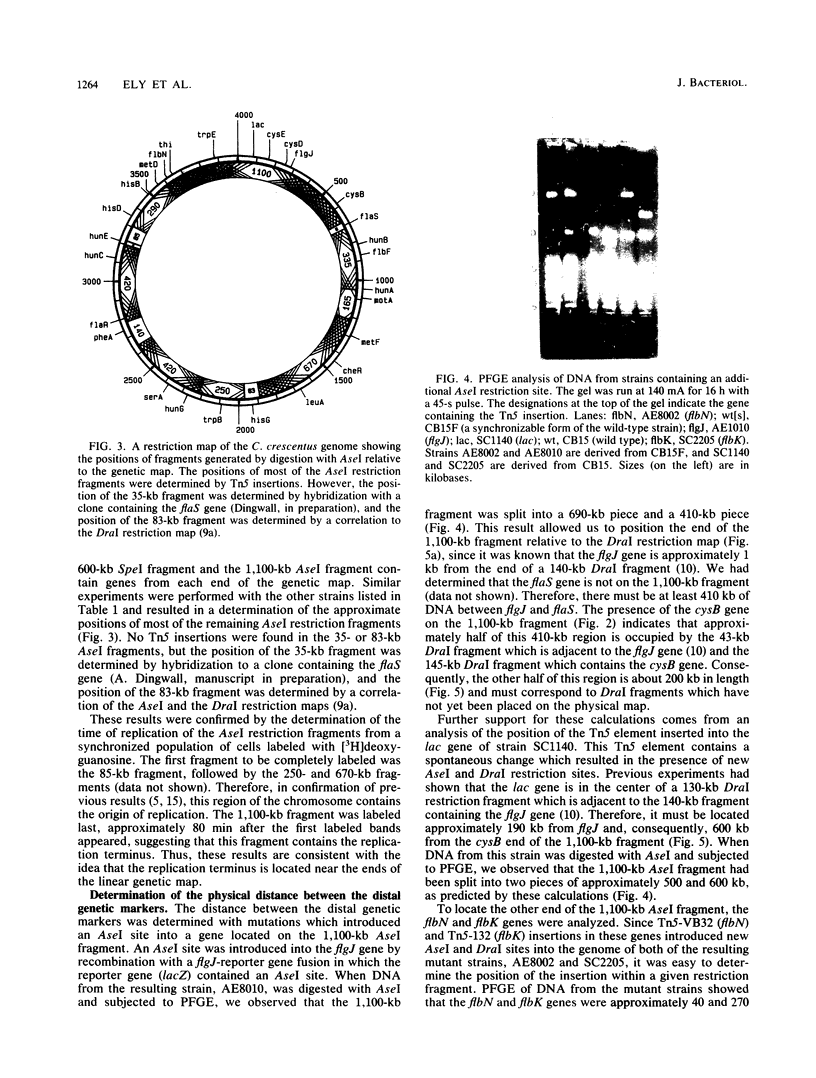
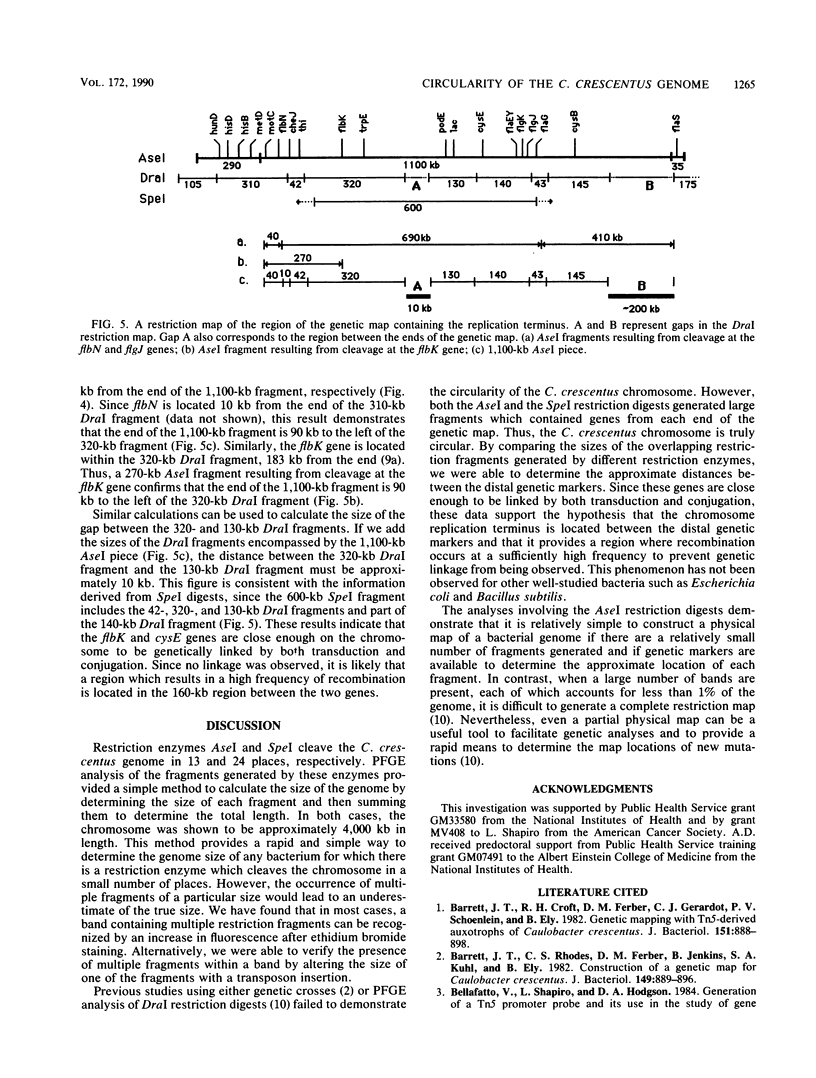

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett J. T., Croft R. H., Ferber D. M., Gerardot C. J., Schoenlein P. V., Ely B. Genetic mapping with Tn5-derived auxotrophs of Caulobacter crescentus. J Bacteriol. 1982 Aug;151(2):888–898. doi: 10.1128/jb.151.2.888-898.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. T., Rhodes C. S., Ferber D. M., Jenkins B., Kuhl S. A., Ely B. Construction of a genetic map for Caulobacter crescentus. J Bacteriol. 1982 Mar;149(3):889–896. doi: 10.1128/jb.149.3.889-896.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellofatto V., Shapiro L., Hodgson D. A. Generation of a Tn5 promoter probe and its use in the study of gene expression in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1035–1039. doi: 10.1073/pnas.81.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champer R., Dingwall A., Shapiro L. Cascade regulation of Caulobacter flagellar and chemotaxis genes. J Mol Biol. 1987 Mar 5;194(1):71–80. doi: 10.1016/0022-2836(87)90716-9. [DOI] [PubMed] [Google Scholar]
- Dingwall A., Shapiro L. Rate, origin, and bidirectionality of Caulobacter chromosome replication as determined by pulsed-field gel electrophoresis. Proc Natl Acad Sci U S A. 1989 Jan;86(1):119–123. doi: 10.1073/pnas.86.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H. Transposon mutagenesis in Caulobacter crescentus. J Bacteriol. 1982 Feb;149(2):620–625. doi: 10.1128/jb.149.2.620-625.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Ely T. W. Use of pulsed field gel electrophoresis and transposon mutagenesis to estimate the minimal number of genes required for motility in Caulobacter crescentus. Genetics. 1989 Dec;123(4):649–654. doi: 10.1093/genetics/123.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Gerardot C. J. Use of pulsed-field-gradient gel electrophoresis to construct a physical map of the Caulobacter crescentus genome. Gene. 1988 Sep 7;68(2):323–333. doi: 10.1016/0378-1119(88)90035-2. [DOI] [PubMed] [Google Scholar]
- Ely B., Johnson R. C. Generalized Transduction in CAULOBACTER CRESCENTUS. Genetics. 1977 Nov;87(3):391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Transfer of drug resistance factors to the dimorphic bacterium Caulobacter crescentus. Genetics. 1979 Mar;91(3):371–380. doi: 10.1093/genetics/91.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Vectors for transposon mutagenesis of non-enteric bacteria. Mol Gen Genet. 1985;200(2):302–304. doi: 10.1007/BF00425440. [DOI] [PubMed] [Google Scholar]
- Evinger M., Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977 Oct;132(1):294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdows M. S., Barbour A. G. Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5969–5973. doi: 10.1073/pnas.86.15.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J Bacteriol. 1979 Jan;137(1):627–634. doi: 10.1128/jb.137.1.627-634.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott T., Ohta N., Newton A. Order of gene replication in Caulobacter crescentus; use of in vivo labeled genomic DNA as a probe. Mol Gen Genet. 1987 Dec;210(3):543–550. doi: 10.1007/BF00327210. [DOI] [PubMed] [Google Scholar]