Abstract
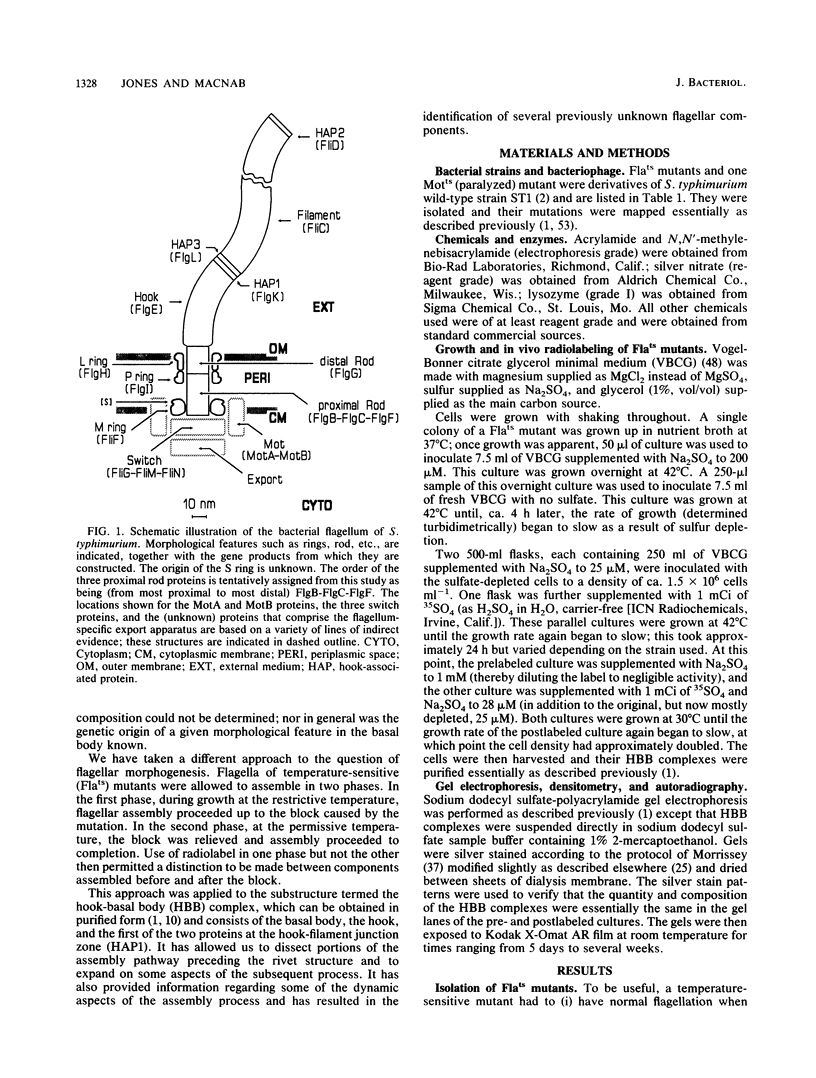
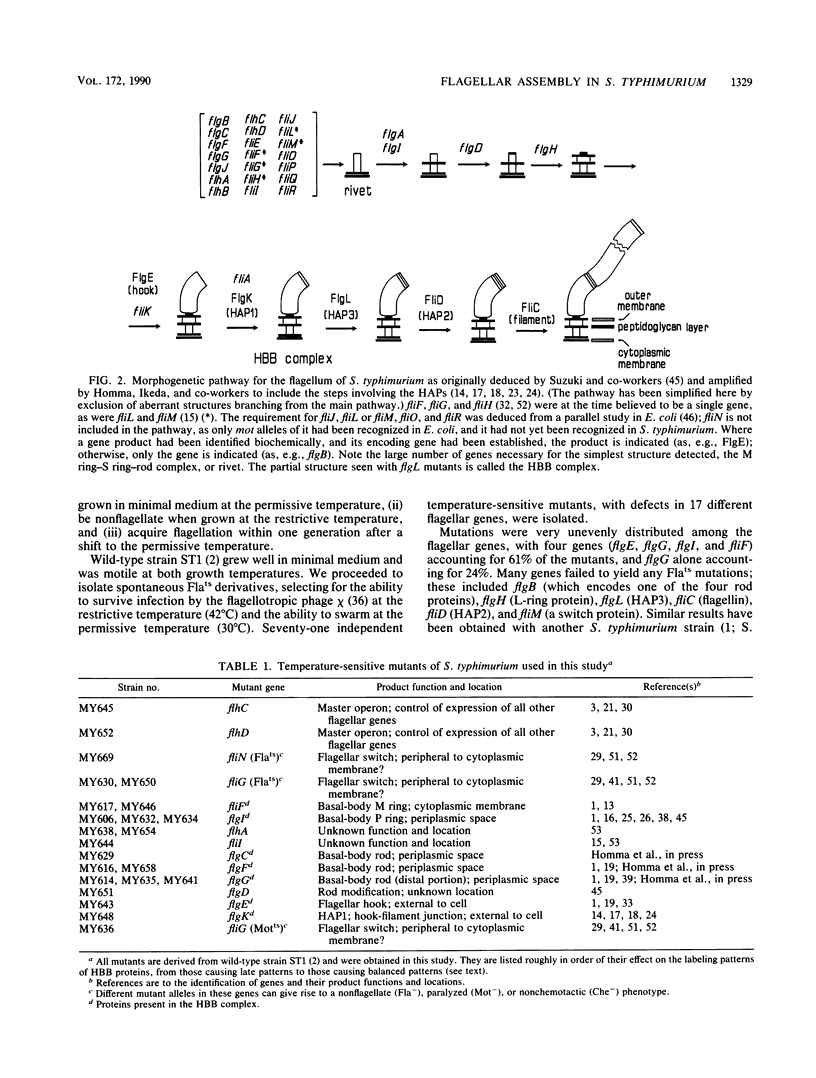
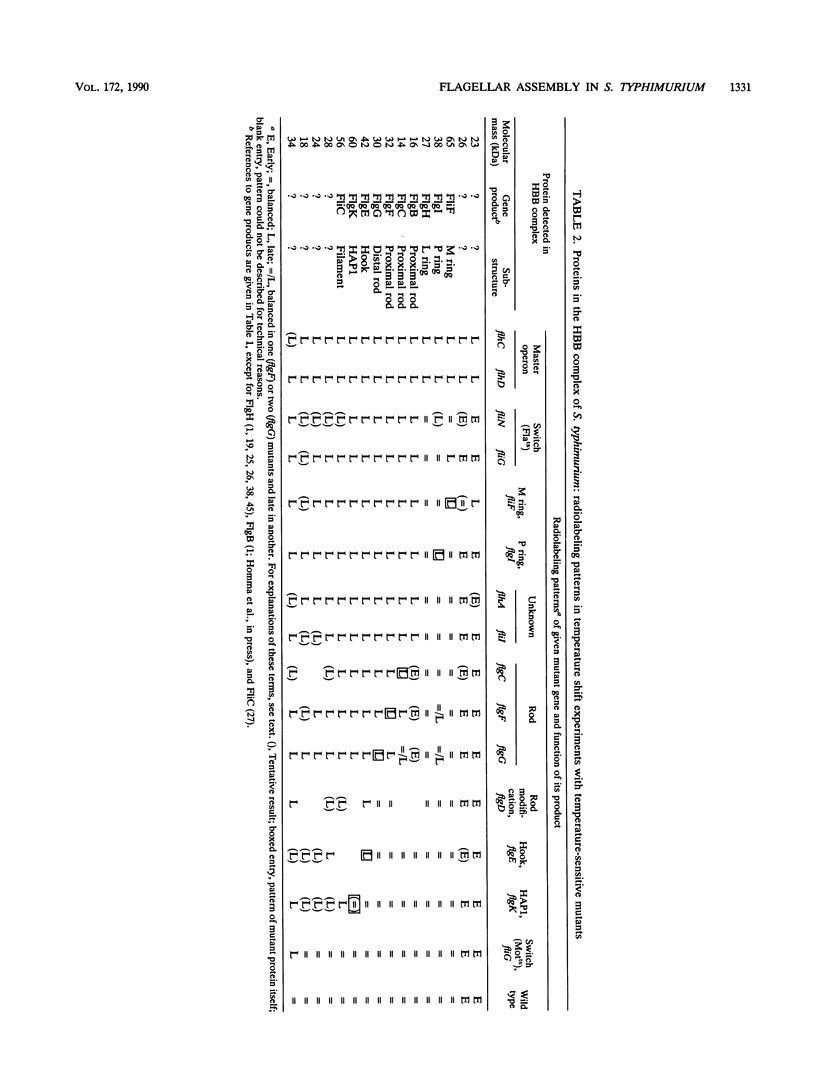
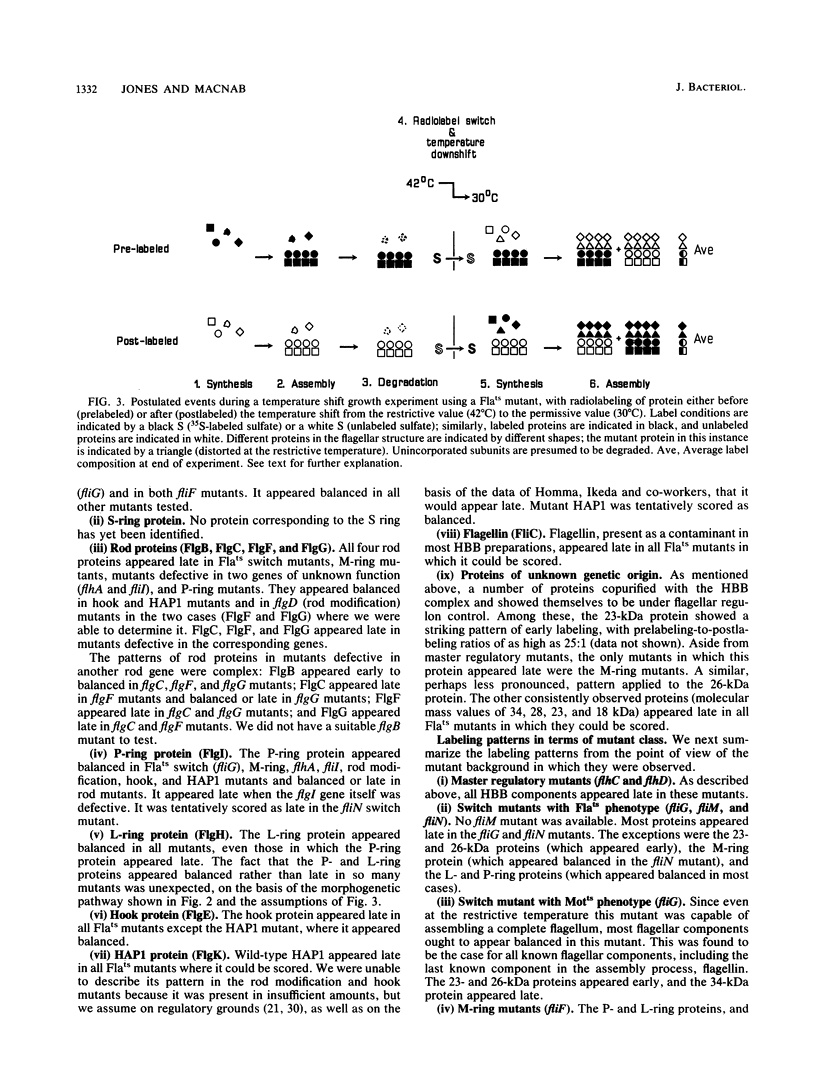
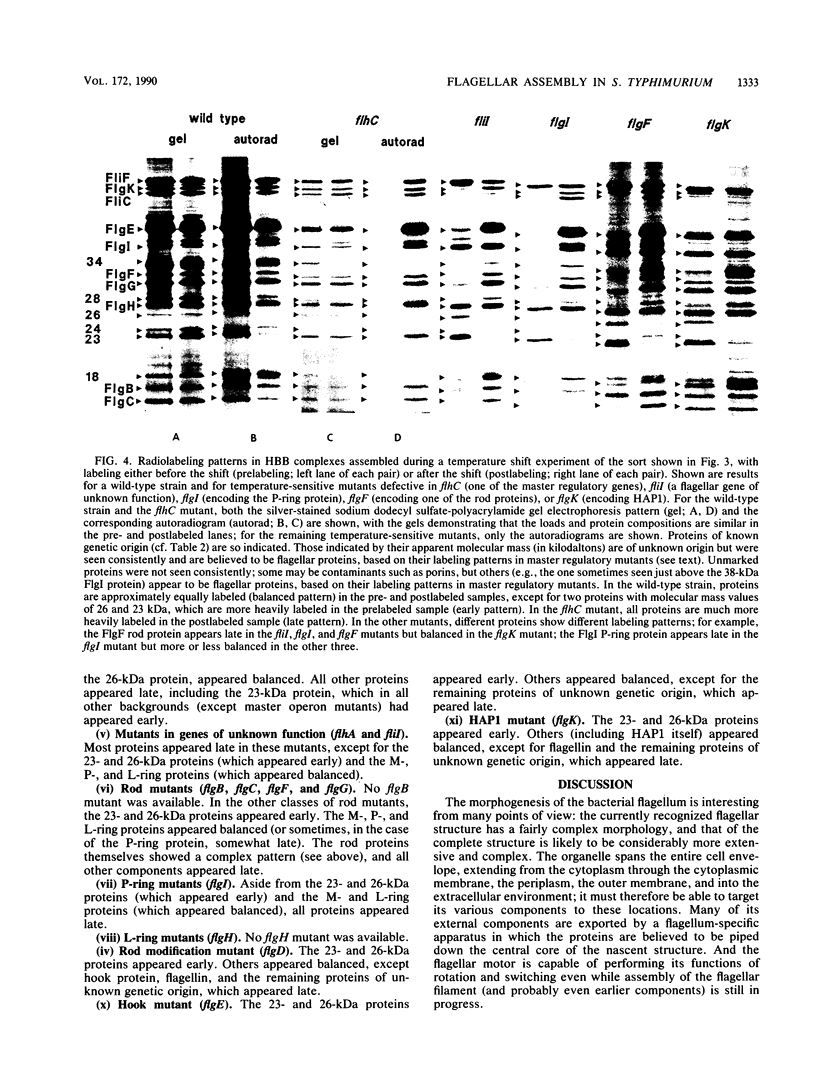
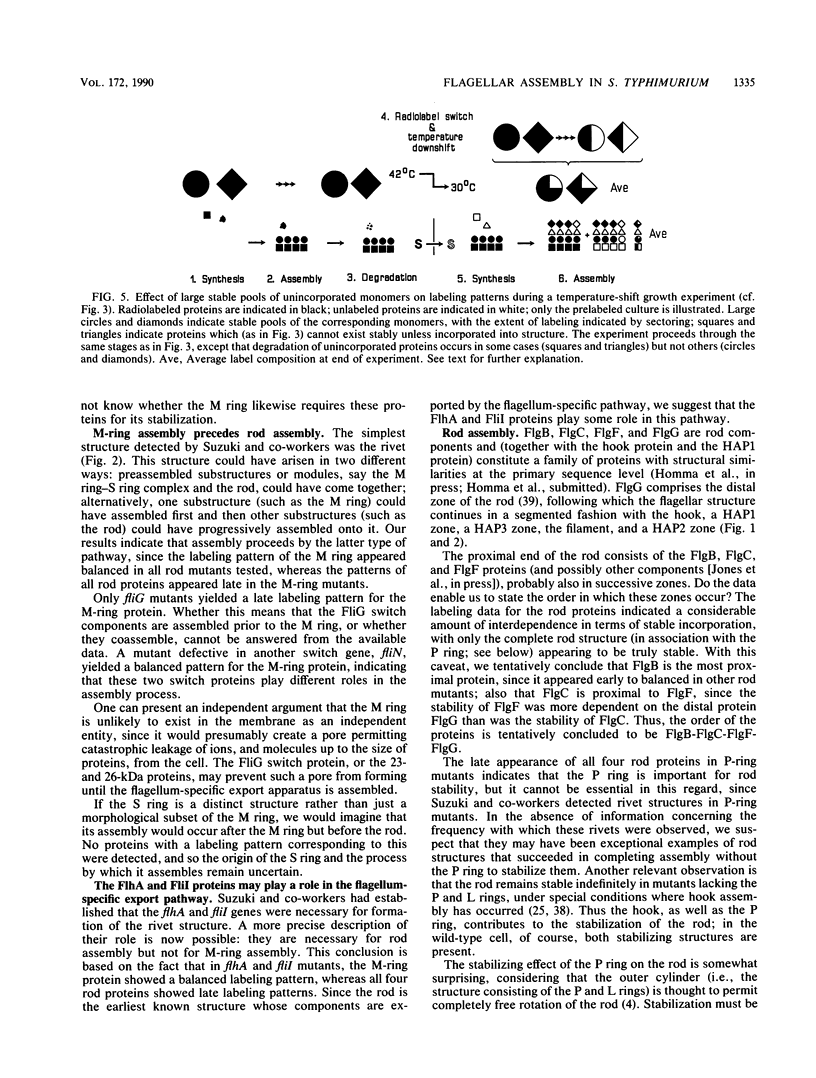
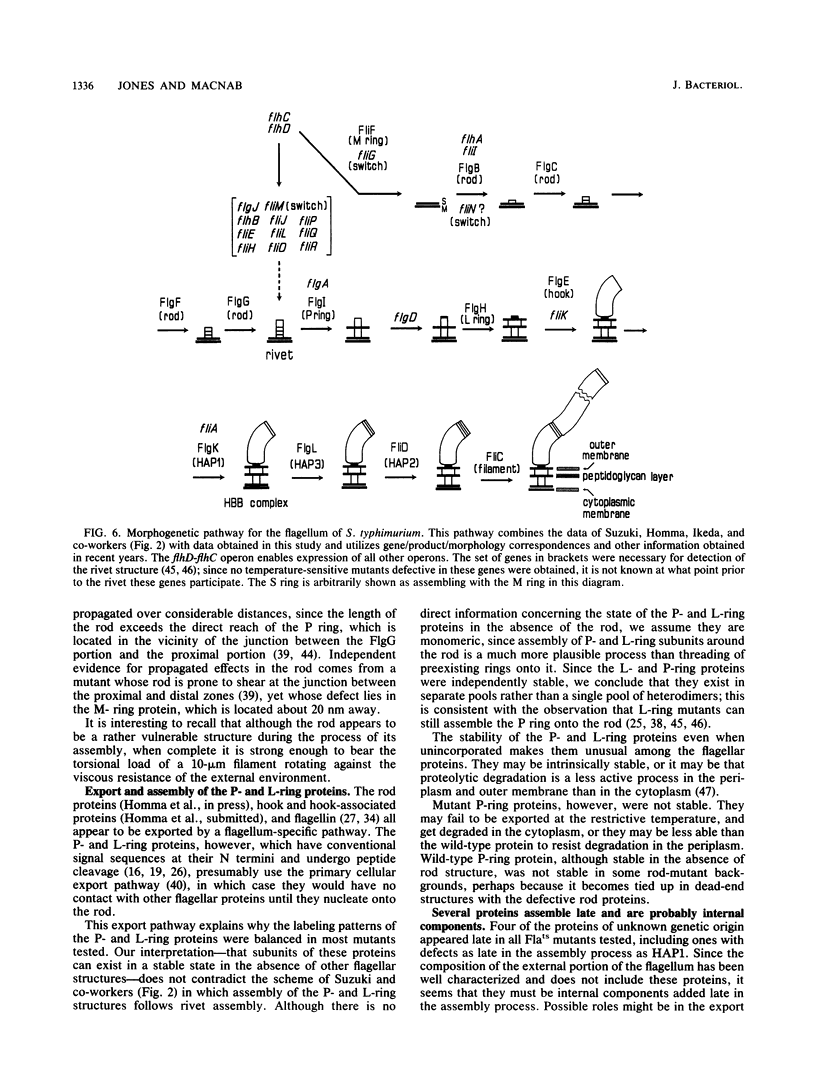
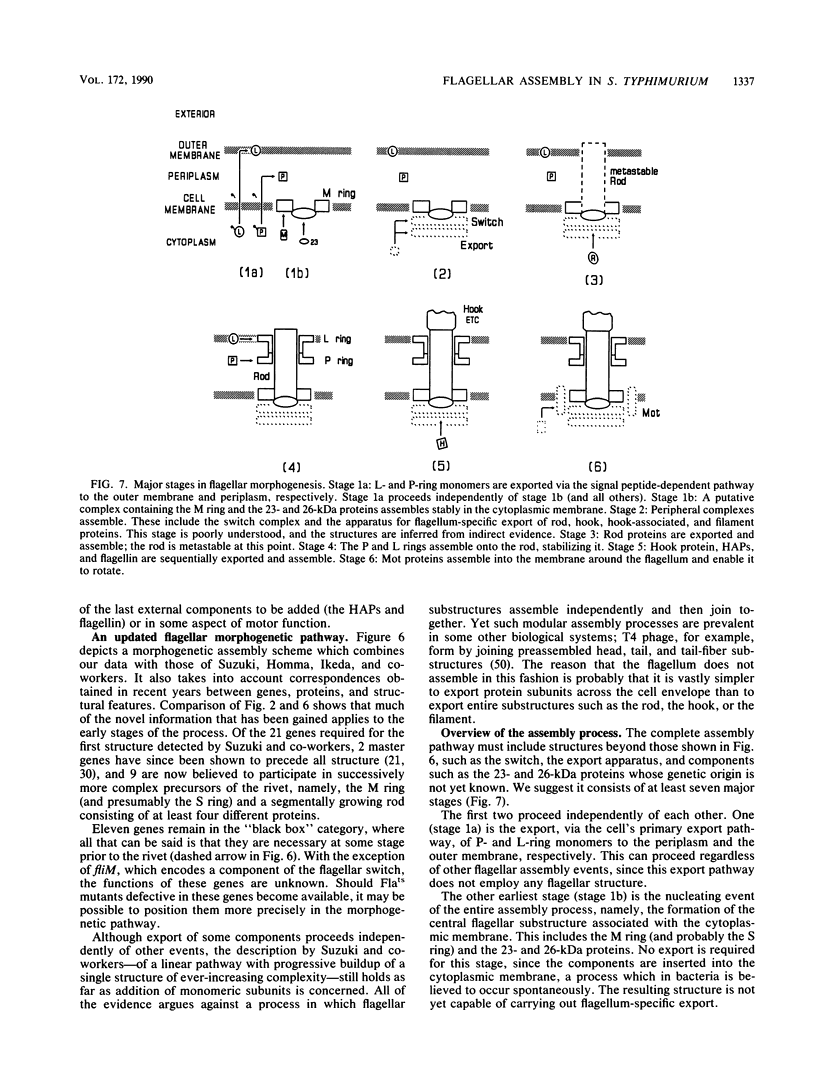
The process of flagellar assembly in Salmonella typhimurium was investigated by using temperature-sensitive mutants. The mutants were grown at the restrictive temperature and then at the permissive temperature, with radiolabel supplied in the first phase of the experiment and not the second, or vice versa. Flagellar hook-basal body complexes were then purified and analyzed by gel electrophoresis and autoradiography. The extent to which a given protein was labeled in the two phases of the experiment provided information as to whether it preceded or followed the block caused by the mutant protein. We conclude the following concerning flagellar assembly. The M-ring protein (FliF) is stably incorporated in the earliest stage detected, along with two previously unknown proteins, with apparent molecular masses of 23 and 26 kilodaltons, respectively, and possibly one of the switch components, FliG. Independent of that event and all other events, the P-ring and L-ring proteins (FlgI and FlgH) are synthesized and exported to the periplasm and outer membrane by the primary cellular export pathway. Rod assembly occurs by export (via the flagellum-specific pathway) of subunits of four proteins, FlgB, FlgC, FlgF, and FlgG, and their incorporation, probably in that order, into the rod structure; this stage requires the flhA and fliI genes, perhaps because they encode part of the export apparatus. Once rod assembly is complete, the FlgI and FlgH proteins assemble around the rod to form the P and L rings. The rod structure, which is only metastable while it is being constructed, becomes stable upon P-ring addition. Export (via the flagellum-specific pathway) and assembly of hook protein, hook-associated proteins, and filament protein then occur successively. A number of flagellar proteins, whose genetic origin and structural role are not yet known, were identified on the basis of their dependence on the flagellar master operon for expression.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aizawa S. I., Dean G. E., Jones C. J., Macnab R. M., Yamaguchi S. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J Bacteriol. 1985 Mar;161(3):836–849. doi: 10.1128/jb.161.3.836-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswad D., Koshland D. E., Jr Isolation, characterization and complementation of Salmonella typhimurium chemotaxis mutants. J Mol Biol. 1975 Sep 15;97(2):225–235. doi: 10.1016/s0022-2836(75)80036-2. [DOI] [PubMed] [Google Scholar]
- Bartlett D. H., Frantz B. B., Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5' consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988 Apr;170(4):1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H. C. Dynamic properties of bacterial flagellar motors. Nature. 1974 May 3;249(452):77–79. doi: 10.1038/249077a0. [DOI] [PubMed] [Google Scholar]
- Berg H. C., Manson M. D., Conley M. P. Dynamics and energetics of flagellar rotation in bacteria. Symp Soc Exp Biol. 1982;35:1–31. [PubMed] [Google Scholar]
- Blair D. F., Berg H. C. Restoration of torque in defective flagellar motors. Science. 1988 Dec 23;242(4886):1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- Block S. M., Berg H. C. Successive incorporation of force-generating units in the bacterial rotary motor. 1984 May 31-Jun 6Nature. 309(5967):470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- Chun S. Y., Parkinson J. S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988 Jan 15;239(4837):276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L., Adler J. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J Bacteriol. 1971 Jan;105(1):384–395. doi: 10.1128/jb.105.1.384-395.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L., Adler J. Purification of intact flagella from Escherichia coli and Bacillus subtilis. J Bacteriol. 1971 Jan;105(1):376–383. doi: 10.1128/jb.105.1.376-383.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G. E., Macnab R. M., Stader J., Matsumura P., Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984 Sep;159(3):991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. U., Tokuyasu K., Simon M. I. Bacterial flagella: polarity of elongation. Science. 1970 Jul 10;169(3941):190–192. doi: 10.1126/science.169.3941.190. [DOI] [PubMed] [Google Scholar]
- Homma M., Aizawa S., Dean G. E., Macnab R. M. Identification of the M-ring protein of the flagellar motor of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7483–7487. doi: 10.1073/pnas.84.21.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Iino T. Locations of hook-associated proteins in flagellar structures of Salmonella typhimurium. J Bacteriol. 1985 Apr;162(1):183–189. doi: 10.1128/jb.162.1.183-189.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Iino T., Macnab R. M. Identification and characterization of the products of six region III flagellar genes (flaAII.3 through flaQII) of Salmonella typhimurium. J Bacteriol. 1988 May;170(5):2221–2228. doi: 10.1128/jb.170.5.2221-2228.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Komeda Y., Iino T., Macnab R. M. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol. 1987 Apr;169(4):1493–1498. doi: 10.1128/jb.169.4.1493-1498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Iino T. Structural genes for flagellar hook-associated proteins in Salmonella typhimurium. J Bacteriol. 1985 Aug;163(2):464–471. doi: 10.1128/jb.163.2.464-471.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Iino T., Yamaguchi S. Hook-associated proteins essential for flagellar filament formation in Salmonella typhimurium. J Bacteriol. 1984 Jan;157(1):100–108. doi: 10.1128/jb.157.1.100-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Ohnishi K., Iino T., Macnab R. M. Identification of flagellar hook and basal body gene products (FlaFV, FlaFVI, FlaFVII and FlaFVIII) in Salmonella typhimurium. J Bacteriol. 1987 Aug;169(8):3617–3624. doi: 10.1128/jb.169.8.3617-3624.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T., Komeda Y., Kutsukake K., Macnab R. M., Matsumura P., Parkinson J. S., Simon M. I., Yamaguchi S. New unified nomenclature for the flagellar genes of Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1988 Dec;52(4):533–535. doi: 10.1128/mr.52.4.533-535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T. Polarity of flagellar growth in salmonella. J Gen Microbiol. 1969 May;56(2):227–239. doi: 10.1099/00221287-56-2-227. [DOI] [PubMed] [Google Scholar]
- Ikeda T., Asakura S., Kamiya R. "Cap" on the tip of Salmonella flagella. J Mol Biol. 1985 Aug 20;184(4):735–737. doi: 10.1016/0022-2836(85)90317-1. [DOI] [PubMed] [Google Scholar]
- Ikeda T., Homma M., Iino T., Asakura S., Kamiya R. Localization and stoichiometry of hook-associated proteins within Salmonella typhimurium flagella. J Bacteriol. 1987 Mar;169(3):1168–1173. doi: 10.1128/jb.169.3.1168-1173.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Homma M., Macnab R. M. Identification of proteins of the outer (L and P) rings of the flagellar basal body of Escherichia coli. J Bacteriol. 1987 Apr;169(4):1489–1492. doi: 10.1128/jb.169.4.1489-1492.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Homma M., Macnab R. M. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jul;171(7):3890–3900. doi: 10.1128/jb.171.7.3890-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joys T. M. The covalent structure of the phase-1 flagellar filament protein of Salmonella typhimurium and its comparison with other flagellins. J Biol Chem. 1985 Dec 15;260(29):15758–15761. [PubMed] [Google Scholar]
- Khan S., Dapice M., Reese T. S. Effects of mot gene expression on the structure of the flagellar motor. J Mol Biol. 1988 Aug 5;202(3):575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- Kihara M., Homma M., Kutsukake K., Macnab R. M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jun;171(6):3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Fusions of flagellar operons to lactose genes on a mu lac bacteriophage. J Bacteriol. 1982 Apr;150(1):16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y., Silverman M., Matsumura P., Simon M. Genes for the hook-basal body proteins of the flagellar apparatus in Escherichia coli. J Bacteriol. 1978 May;134(2):655–667. doi: 10.1128/jb.134.2.655-667.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K., Iino T., Komeda Y., Yamaguchi S. Functional homology of fla genes between Salmonella typhimurium and Escherichia coli. Mol Gen Genet. 1980 Apr;178(1):59–67. doi: 10.1007/BF00267213. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Suzuki T., Yamaguchi S., Iino T. Role of gene flaFV on flagellar hook formation in Salmonella typhimurium. J Bacteriol. 1979 Oct;140(1):267–275. doi: 10.1128/jb.140.1.267-275.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima G., Kawagishi I., Homma M., Asaka J., Kondo E., Macnab R. M. Export of an N-terminal fragment of Escherichia coli flagellin by a flagellum-specific pathway. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4953–4957. doi: 10.1073/pnas.86.13.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEYNELL E. W. A phage, phi chi, which attacks motile bacteria. J Gen Microbiol. 1961 Jun;25:253–290. doi: 10.1099/00221287-25-2-253. [DOI] [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Homma M., Kutsukake K., Iino T. Formation of flagella lacking outer rings by flaM, flaU, and flaY mutants of Escherichia coli. J Bacteriol. 1987 Apr;169(4):1485–1488. doi: 10.1128/jb.169.4.1485-1488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okino H., Isomura M., Yamaguchi S., Magariyama Y., Kudo S., Aizawa S. I. Release of flagellar filament-hook-rod complex by a Salmonella typhimurium mutant defective in the M ring of the basal body. J Bacteriol. 1989 Apr;171(4):2075–2082. doi: 10.1128/jb.171.4.2075-2082.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- Ravid S., Eisenbach M. Direction of flagellar rotation in bacterial cell envelopes. J Bacteriol. 1984 Apr;158(1):222–230. doi: 10.1128/jb.158.1.222-230.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Matsumura P., Simon M. The identification of the mot gene product with Escherichia coli-lambda hybrids. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3126–3130. doi: 10.1073/pnas.73.9.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stader J., Matsumura P., Vacante D., Dean G. E., Macnab R. M. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986 Apr;166(1):244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallmeyer M. J., Aizawa S., Macnab R. M., DeRosier D. J. Image reconstruction of the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1989 Feb 5;205(3):519–528. doi: 10.1016/0022-2836(89)90223-4. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Iino T., Horiguchi T., Yamaguchi S. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J Bacteriol. 1978 Feb;133(2):904–915. doi: 10.1128/jb.133.2.904-915.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Komeda Y. Incomplete flagellar structures in Escherichia coli mutants. J Bacteriol. 1981 Feb;145(2):1036–1041. doi: 10.1128/jb.145.2.1036-1041.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge K., Gilbert W. Cellular location affects protein stability in Escherichia coli. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1830–1833. doi: 10.1073/pnas.79.6.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Wilson M. L., Macnab R. M. Overproduction of the MotA protein of Escherichia coli and estimation of its wild-type level. J Bacteriol. 1988 Feb;170(2):588–597. doi: 10.1128/jb.170.2.588-597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Aizawa S., Kihara M., Isomura M., Jones C. J., Macnab R. M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986 Dec;168(3):1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Fujita H., Ishihara A., Aizawa S., Macnab R. M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986 Apr;166(1):187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Iino T., Horiguchi T., Ota K. Genetic analysis of fla and mot cistrons closely linked to H1 in Salmonella abortusequi and its derivatives. J Gen Microbiol. 1972 Apr;70(1):59–75. doi: 10.1099/00221287-70-1-59. [DOI] [PubMed] [Google Scholar]