Abstract
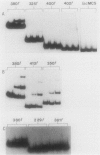
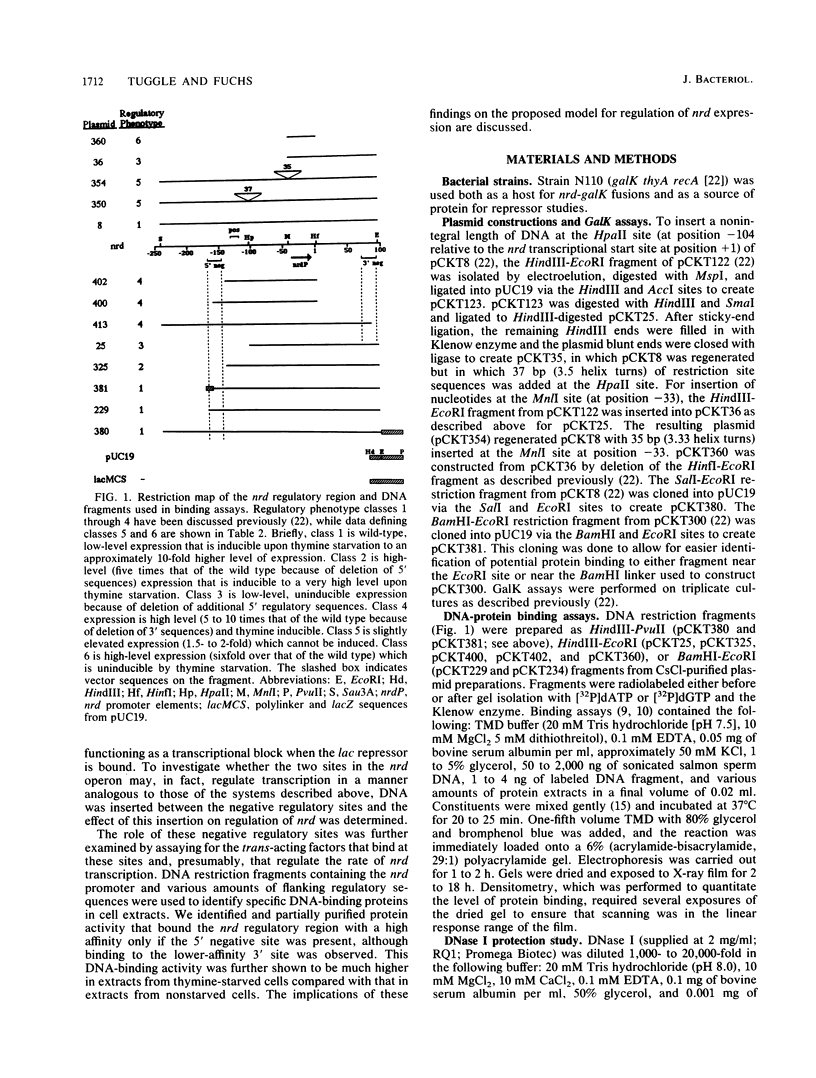
Expression of the nrd genes was previously shown to be controlled by both positive and negative regulation (C. K. Tuggle and J. A. Fuchs, EMBO J. 5:1077-1085, 1986). Two regions, one located 5' and one located 3' of the nrd promoter (nrdP), were identified as negative regulatory sites since deletion of these sequences increased nrd expression. These regions of DNA have sequence similarities, and a looping mechanism was proposed to explain the requirement for two distinct sites in nrd repression. To investigate the role of these sequences in regulating nrd, a gel electrophoresis assay was used to detect the proteins that bind to the nrd regulatory sites. A protein that bound to restriction fragments containing the negative regulatory sites but not to other DNA fragments was identified in cell extracts and was partially purified. DNase I footprinting experiments showed that the binding protein protects the 5' negative site previously identified in vivo. The 3' negative site also identified in vivo was not required in vitro for high-affinity protein binding to the 5' site, but lower-affinity binding to this site could be detected. Specific binding to the 5' site was found to be elevated approximately 10-fold in crude extracts from thymine-starved cells as compared with that in extracts from unstarved cells. This higher activity was also evident in purified preparations, suggesting that thymine starvation increases the expression of the negative regulatory protein. The finding that a purified protein preparation binds both negative regulatory sites indicates that this preparation contains the nrd repressor protein or proteins. Insertion of 37 base pairs (3.5 helix turns) of DNA at a HpaII site or 35 base pairs (3.3 turns) at a MnlI site between the 5' regulatory sites and nrdP abolished the increase in nrd expression resulting from thymine starvation in vivo, but negative regulation appeared to be less affected than when either negative site was deleted. Insertion of DNA in these constructs was shown not to affect repressor binding in vitro, indicating either that a simple model of DNA looping to bring equivalent operator sites into physical proximity does not explain repression at nrd or that the distance between sites is sufficient that helical turns are of little importance.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carlson J., Fuchs J. A., Messing J. Primary structure of the Escherichia coli ribonucleoside diphosphate reductase operon. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4294–4297. doi: 10.1073/pnas.81.14.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle U., Gentz R., Bujard H. lac Repressor blocks transcribing RNA polymerase and terminates transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4134–4137. doi: 10.1073/pnas.83.12.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Sjöberg B. M., Hahne S. Ribonucleoside diphosphate reductase from Escherichia coli. An immunological assay and a novel purification from an overproducing strain lysogenic for phage lambdadnrd. J Biol Chem. 1977 Sep 10;252(17):6132–6138. [PubMed] [Google Scholar]
- Filpula D., Fuchs J. A. Increased synthesis of ribonucleotide reductase after deoxyribonucleic acid inhibition in various species of bacteria. J Bacteriol. 1979 Aug;139(2):694–696. doi: 10.1128/jb.139.2.694-696.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filpula D., Fuchs J. A. Regulation of ribonucleoside diphosphate reductase synthesis in Escherichia coli: increased enzyme synthesis as a result of inhibition of deoxyribonucleic acid synthesis. J Bacteriol. 1977 Apr;130(1):107–113. doi: 10.1128/jb.130.1.107-113.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashner Y., Gralla J. D. Dual mechanism of repression at a distance in the lac operon. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8968–8972. doi: 10.1073/pnas.85.23.8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J. D. Bacterial gene regulation from distant DNA sites. Cell. 1989 Apr 21;57(2):193–195. doi: 10.1016/0092-8674(89)90955-0. [DOI] [PubMed] [Google Scholar]
- Hamilton E. P., Lee N. Three binding sites for AraC protein are required for autoregulation of araC in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1749–1753. doi: 10.1073/pnas.85.6.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke P. D., Fuchs J. A. Regulation of ribonucleoside diphosphate reductase mRNA synthesis in Escherichia coli. J Bacteriol. 1983 Jun;154(3):1040–1045. doi: 10.1128/jb.154.3.1040-1045.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke P. D., Fuchs J. A. Requirement of protein synthesis for the induction of ribonucleoside diphosphate reductase mRNA in Escherichia coli. Mol Gen Genet. 1984;193(2):327–331. doi: 10.1007/BF00330689. [DOI] [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. F. Regulation of the Escherichia coli L-arabinose operon studied by gel electrophoresis DNA binding assay. J Mol Biol. 1984 Sep 25;178(3):611–628. doi: 10.1016/0022-2836(84)90241-9. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Ishii S., Salstrom J. S., Sugino Y., Szybalski W., Imamoto F. A biochemical assay for the transcription-antitermination function of the coliphage lambda N gene product. Gene. 1980 Jun;10(1):17–25. doi: 10.1016/0378-1119(80)90139-0. [DOI] [PubMed] [Google Scholar]
- Platz A., Sjöberg B. M. Construction and characterization of hybrid plasmids containing the Escherichia coli nrd region. J Bacteriol. 1980 Aug;143(2):561–568. doi: 10.1128/jb.143.2.561-568.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander L., Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- Tuggle C. K., Fuchs J. A. Regulation of the operon encoding ribonucleotide reductase in Escherichia coli: evidence for both positive and negative control. EMBO J. 1986 May;5(5):1077–1085. doi: 10.1002/j.1460-2075.1986.tb04325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]