Abstract
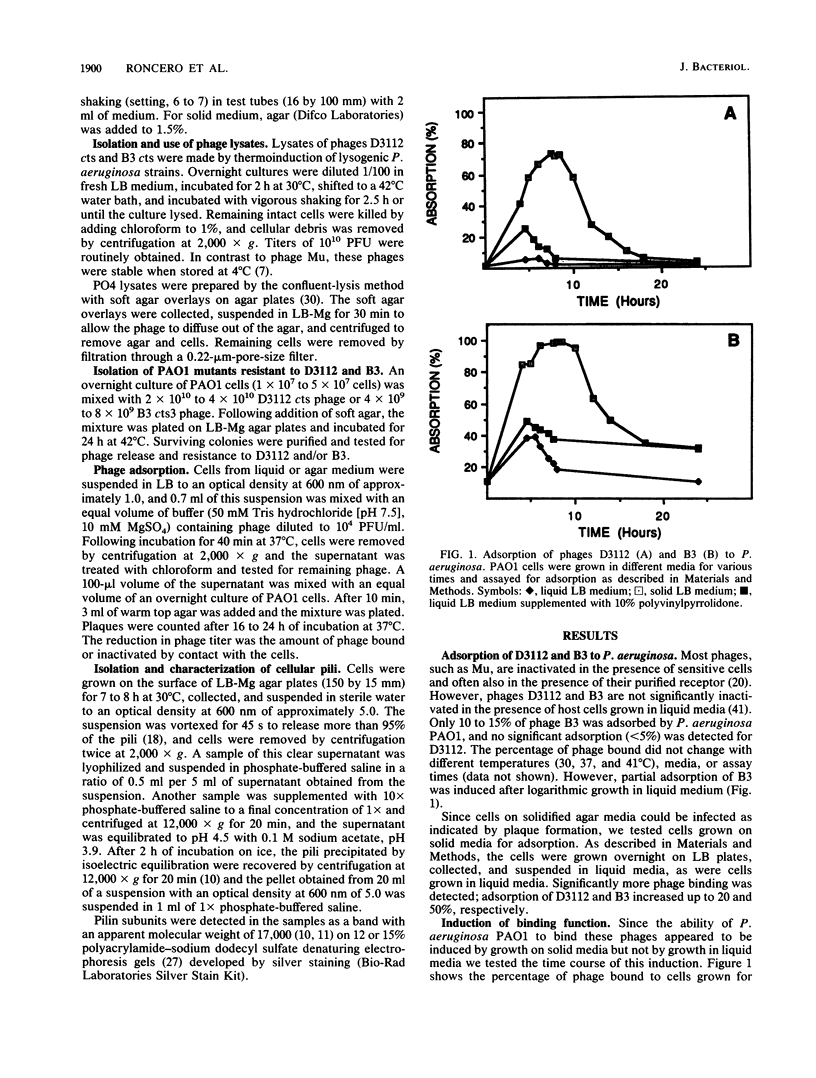
Pseudomonas aeruginosa transposable bacteriophages D3112 and B3 were found to require pili for infection. Seventy mutants of P. aeruginosa PAO selected by resistance to D3112 or B3 were also resistant to the phage not used in the selection and suggested that the receptors of these two phages are identical. Of five resistant mutants examined, all were defective in the production of pili and did not adsorb either phage. P. aeruginosa PAK strains altered in pilus expression, such as hyperpiliated or nonpiliated mutants, adsorbed the phage but were not productively infected, implying that an additional host function was required for infection. The cell-associated lipopolysaccharide was not required for D3112 or B3 infection, since mutants deficient in O side-chain and core biosynthesis were still capable of adsorption and productive infection. This is in contrast to Escherichia coli mutator phages Mu and D108, which are dependent on lipopolysaccharide for adsorption. The P. aeruginosa phages adsorbed only to cells grown on solid media or in liquid media supplemented with agents that increase the macroviscosity, such as polyvinylpyrrolidone. Adsorption time course studies of D3112 and B3 using cells grown in solid media revealed similar but not identical adsorption patterns. These studies suggested that expression of the D3112 and B3 cell receptor is induced by growth on solid media.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akhverdian V. Z., Khrenova E. A., Bogush V. G., Gerasimova T. V., Kirsanov N. B. Shirokaia rasprostranennost' transpozonnykh fagov v prirodnykh populiatsiiakh Pseudomonas aeruginosa. Genetika. 1984 Oct;20(10):1612–1619. [PubMed] [Google Scholar]
- Belas R., Simon M., Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986 Jul;167(1):210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D. E. A pilus-dependent Pseudomonas aeruginosa bacteriophage with a long noncontractile tail. Virology. 1973 Feb;51(2):489–492. doi: 10.1016/0042-6822(73)90448-0. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology. 1974 Mar;58(1):149–163. doi: 10.1016/0042-6822(74)90150-0. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. The adsorption of the Pseudomonas aeruginosa filamentous bacteriophage Pf to its host. Can J Microbiol. 1973 May;19(5):623–631. doi: 10.1139/m73-103. [DOI] [PubMed] [Google Scholar]
- Darzins A., Casadaban M. J. In vivo cloning of Pseudomonas aeruginosa genes with mini-D3112 transposable bacteriophage. J Bacteriol. 1989 Jul;171(7):3917–3925. doi: 10.1128/jb.171.7.3917-3925.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A., Casadaban M. J. Mini-D3112 bacteriophage transposable elements for genetic analysis of Pseudomonas aeruginosa. J Bacteriol. 1989 Jul;171(7):3909–3916. doi: 10.1128/jb.171.7.3909-3916.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman T. C., Hoyne P. A., Stewart D. J., McKern N. M., Peterson J. E. Expression of pili from Bacteroides nodosus in Pseudomonas aeruginosa. J Bacteriol. 1986 Nov;168(2):574–580. doi: 10.1128/jb.168.2.574-580.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Paranchych W. Composition and molecular weight of pili purified from Pseudomonas aeruginosa K. J Bacteriol. 1977 Jul;131(1):259–269. doi: 10.1128/jb.131.1.259-269.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A., Casadaban M. J. Mini-mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusing. J Bacteriol. 1986 Oct;168(1):357–364. doi: 10.1128/jb.168.1.357-364.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLLOWAY B. W., EGAN J. B., MONK M. Lysogeny in Pseudomonas aeruginosa. Aust J Exp Biol Med Sci. 1960 Aug;38:321–329. doi: 10.1038/icb.1960.34. [DOI] [PubMed] [Google Scholar]
- HOLLOWAY B. W. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955 Dec;13(3):572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- Ianenko A. S., Bogush V. G., Kirsanov N. B., Liapin M. N., Krylov V. N. Ispol'zovanie deletsionnykh mutantov plazmidy RP::4D3112 dlia geneticheskogo analiza bakteriofaga D3112 Pseudomonas aeruginosa. Genetika. 1983 Nov;19(11):1760–1768. [PubMed] [Google Scholar]
- Ishimoto K. S., Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Lory S. Characterization of Pseudomonas aeruginosa mutants with altered piliation. J Bacteriol. 1987 Dec;169(12):5663–5667. doi: 10.1128/jb.169.12.5663-5667.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Parker M. L., Lory S. Nucleotide sequence and transcriptional initiation site of two Pseudomonas aeruginosa pilin genes. J Biol Chem. 1986 Nov 25;261(33):15703–15708. [PubMed] [Google Scholar]
- Kropinski A. M., Chan L. C., Milazzo F. H. The extraction and analysis of lipopolysaccharides from Pseudomonas aeruginosa strain PAO, and three rough mutants. Can J Microbiol. 1979 Mar;25(3):390–398. doi: 10.1139/m79-060. [DOI] [PubMed] [Google Scholar]
- Kropinski A. M., Chan L., Jarrell K., Milazzo F. H. The nature of Pseudomonas aeruginosa strain PAO bacteriophage receptors. Can J Microbiol. 1977 Jun;23(6):653–658. doi: 10.1139/m77-098. [DOI] [PubMed] [Google Scholar]
- Krylov V. N., Bogush V. G., Ianenko A. S., Kirsanov N. B. Bakteriofagi Pseudomonas aeruginosa, struktura DNA kotorykh skhodna so strukturoi DNK faga Mu1. Soobshchenie II. Dokazatel'stvo rodstva bakteriofagov D3112, B3 i B39: analiz rasshchepleniia DNK éndonukleazami restriktsii, vydelenie rekombinanta fagov D3112 i B3. Genetika. 1980;16(6):975–984. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marrs C. F., Schoolnik G., Koomey J. M., Hardy J., Rothbard J., Falkow S. Cloning and sequencing of a Moraxella bovis pilin gene. J Bacteriol. 1985 Jul;163(1):132–139. doi: 10.1128/jb.163.1.132-139.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKern N. M., O'Donnell I. J., Inglis A. S., Stewart D. J., Clark B. L. Amino acid sequence of pilin from Bacteroides nodosus (strain 198), the causative organism of ovine footrot. FEBS Lett. 1983 Nov 28;164(1):149–153. doi: 10.1016/0014-5793(83)80039-8. [DOI] [PubMed] [Google Scholar]
- Morgan T. M., Stanisich V. A. Characterization and properties of phage B33, a female-specific phage of Pseudomonas aeruginosa. J Gen Virol. 1976 Jan;30(1):73–79. doi: 10.1099/0022-1317-30-1-73. [DOI] [PubMed] [Google Scholar]
- Murooka Y., Takizawa N., Harada T. Introduction of bacteriophage Mu into bacteria of various genera and intergeneric gene transfer by RP4::Mu. J Bacteriol. 1981 Jan;145(1):358–368. doi: 10.1128/jb.145.1.358-368.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmat S., Shapiro J. A. Insertion and replication of the Pseudomonas aeruginosa mutator phage D3112. Mol Gen Genet. 1983;192(3):416–423. doi: 10.1007/BF00392184. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Rothbard J. B., Schoolnik G. K. The primary sequence and antigenic structure of gonococcal pilin: approaches towards a gonococcal vaccine. Adv Exp Med Biol. 1985;185:247–273. doi: 10.1007/978-1-4684-7974-4_17. [DOI] [PubMed] [Google Scholar]
- Sastry P. A., Finlay B. B., Pasloske B. L., Paranchych W., Pearlstone J. R., Smillie L. B. Comparative studies of the amino acid and nucleotide sequences of pilin derived from Pseudomonas aeruginosa PAK and PAO. J Bacteriol. 1985 Nov;164(2):571–577. doi: 10.1128/jb.164.2.571-577.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. K., Miller V. L., Furlong D. B., Mekalanos J. J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A. 1987 May;84(9):2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. E., Straus D. C., Johanson W. G., Jr, Berry V. K., Bass J. A. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect Immun. 1980 Sep;29(3):1146–1151. doi: 10.1128/iai.29.3.1146-1151.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]