Abstract
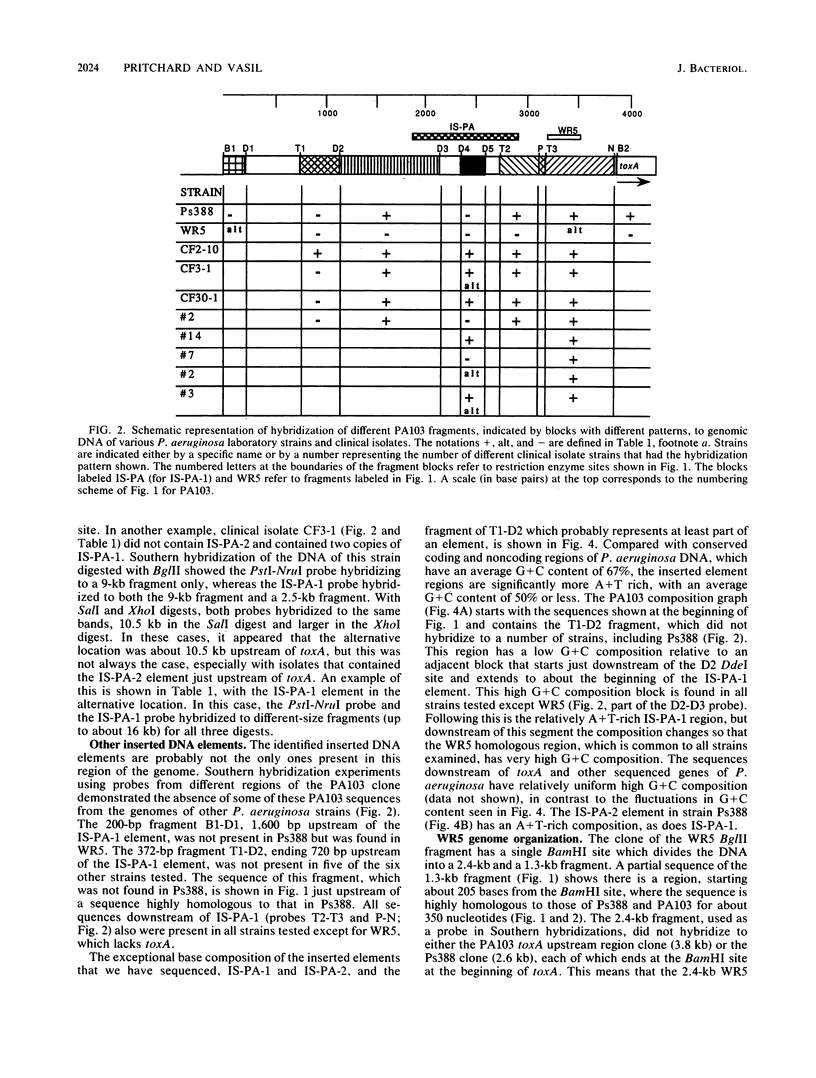
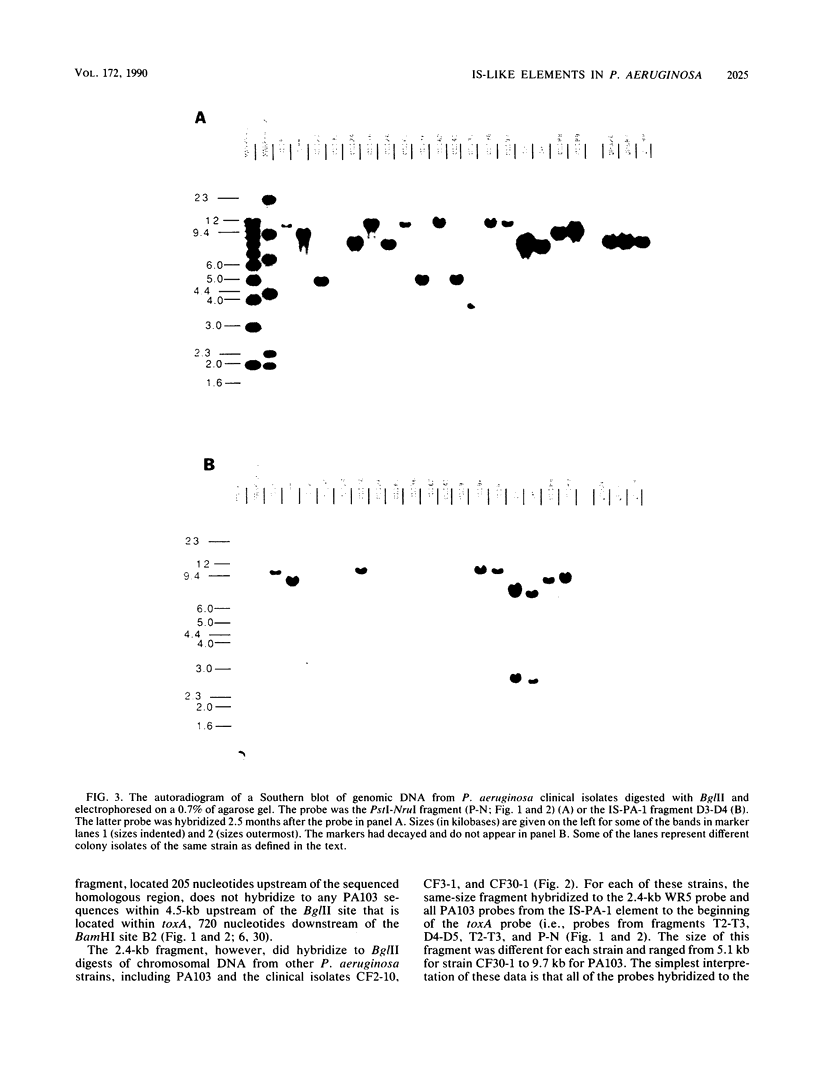
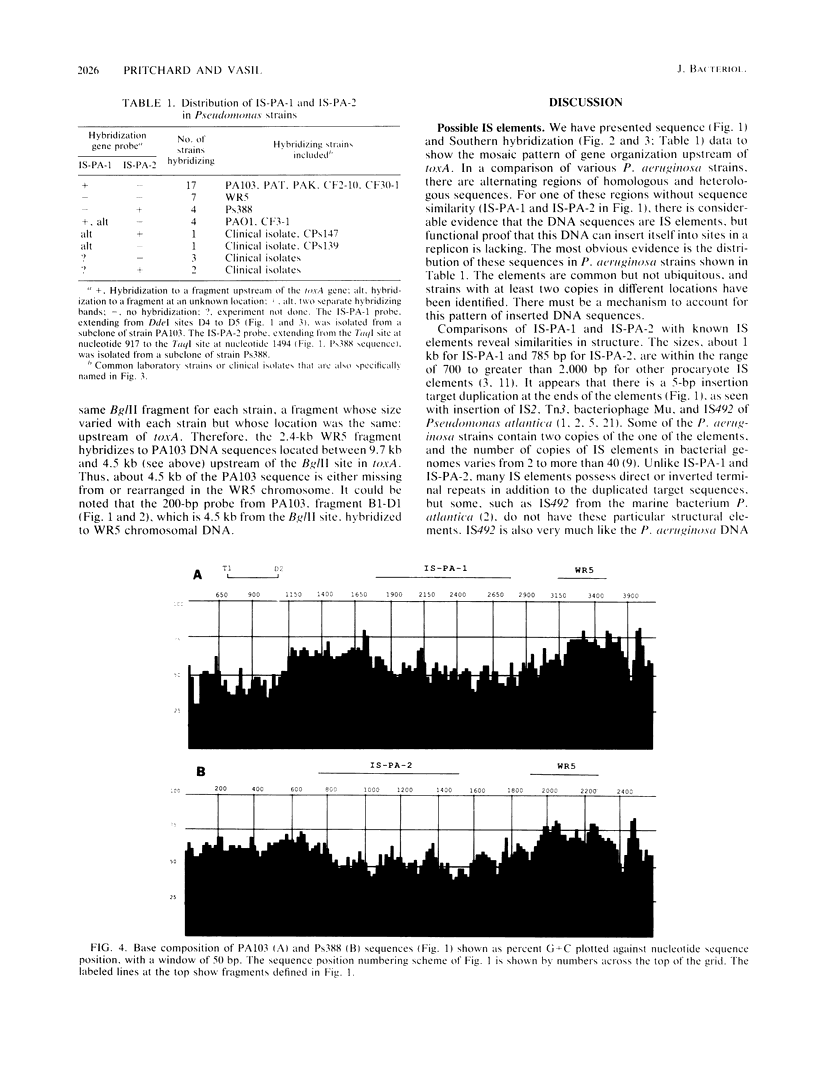
Nucleotide sequence and Southern hybridization data revealed a mosaic genome organization in a region that extends several thousand base pairs upstream of the exotoxin A (toxA) gene in Pseudomonas aeruginosa. An interstrain comparison of DNA in this region showed a pattern of alternating segments of homologous and nonhomologous sequences. Two nonhomologous elements, approximately 1 kilobase pair upstream of the gene in strains PA103 and Ps388, were characterized in more detail. The sequence elements, denoted IS-PA-1 and IS-PA-2 for the different strains, are about 1,000 and 785 base pairs long, respectively, and have 5-base-pair direct repeats at their boundaries, consistent with their being DNA insertion sequences. The distribution of these elements in 34 different strains was determined. IS-PA-1 was found in a single copy upstream of toxA in half of the strains and was found in two copies in four of the strains. Some strains contained neither element, and one strain carried both. The genome of another strain, WR5, which lacks toxA, was shown to contain a 350-base-pair region that was highly homologous to DNA sequences located just upstream of toxA in other strains. The WR5 genome lacked several kilobase pairs of DNA that was found both upstream and downstream of this homologous region in the other strains.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B. Mu insertion duplicates a 5 base pair sequence at the host inserted site. Cell. 1979 Jan;16(1):123–129. doi: 10.1016/0092-8674(79)90193-4. [DOI] [PubMed] [Google Scholar]
- Bartlett D. H., Silverman M. Nucleotide sequence of IS492, a novel insertion sequence causing variation in extracellular polysaccharide production in the marine bacterium Pseudomonas atlantica. J Bacteriol. 1989 Mar;171(3):1763–1766. doi: 10.1128/jb.171.3.1763-1766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Caruso M., Shapiro J. A. Interactions of Tn7 and temperate phage F116L of Pseudomonas aeruginosa. Mol Gen Genet. 1982;188(2):292–298. doi: 10.1007/BF00332690. [DOI] [PubMed] [Google Scholar]
- Danna K. J. Determination of fragment order through partial digests and multiple enzyme digests. Methods Enzymol. 1980;65(1):449–467. doi: 10.1016/s0076-6879(80)65055-1. [DOI] [PubMed] [Google Scholar]
- Ghosal D., Sommer H., Saedler H. Nucleotide sequence of the transposable DNA-element IS2. Nucleic Acids Res. 1979 Mar;6(3):1111–1122. doi: 10.1093/nar/6.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. L., Smith D. H., Baldridge J. S., Harkins R. N., Vasil M. L., Chen E. Y., Heyneker H. L. Cloning, nucleotide sequence, and expression in Escherichia coli of the exotoxin A structural gene of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1984 May;81(9):2645–2649. doi: 10.1073/pnas.81.9.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanne L. F., Howe T. R., Iglewski B. H. Locus of the Pseudomonas aeruginosa toxin A gene. J Bacteriol. 1983 Apr;154(1):383–386. doi: 10.1128/jb.154.1.383-386.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B. W., Morgan A. F. Genome organization in Pseudomonas. Annu Rev Microbiol. 1986;40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- Iyobe S., Sagai H., Mitsuhashi S. Tn2001, a transposon encoding chloramphenicol resistance in Pseudomonas aeruginosa. J Bacteriol. 1981 Apr;146(1):141–148. doi: 10.1128/jb.146.1.141-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- McPheat W. L., Hanson J. H., Livey I., Robertson J. S. Analysis of the chromosomal location of two copies of a Bordetella pertussis insertion sequence. Mol Microbiol. 1989 Jul;3(7):985–989. doi: 10.1111/j.1365-2958.1989.tb00248.x. [DOI] [PubMed] [Google Scholar]
- Mekalanos J. J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983 Nov;35(1):253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- Nash J. H., Krishnapillai V. Identification of an insertion sequence in the chromosome of Pseudomonas aeruginosa PAO. J Bacteriol. 1982 Oct;152(1):514–516. doi: 10.1128/jb.152.1.514-516.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle J. W., Janda J. M., Woods D. E., Vasil M. L. Characterization and use of a DNA probe as an epidemiological marker for Pseudomonas aeruginosa. J Infect Dis. 1987 Jan;155(1):119–126. doi: 10.1093/infdis/155.1.119. [DOI] [PubMed] [Google Scholar]
- Ogle J. W., Reller L. B., Vasil M. L. Development of resistance in Pseudomonas aeruginosa to imipenem, norfloxacin, and ciprofloxacin during therapy: proof provided by typing with a DNA probe. J Infect Dis. 1988 Apr;157(4):743–748. doi: 10.1093/infdis/157.4.743. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohmori H., Ohtsubo E. Nucleotide-sequence analysis of Tn3 (ap): implications for insertion and deletion. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1269–1277. doi: 10.1101/sqb.1979.043.01.144. [DOI] [PubMed] [Google Scholar]
- Pasloske B. L., Joffe A. M., Sun Q., Volpel K., Paranchych W., Eftekhar F., Speert D. P. Serial isolates of Pseudomonas aeruginosa from a cystic fibrosis patient have identical pilin sequences. Infect Immun. 1988 Mar;56(3):665–672. doi: 10.1128/iai.56.3.665-672.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R., Perugini M., Ratti G. DNA element of Corynebacterium diphtheriae with properties of an insertion sequence and usefulness for epidemiological studies. J Bacteriol. 1987 Jan;169(1):308–312. doi: 10.1128/jb.169.1.308-312.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scordilis G. E., Ree H., Lessie T. G. Identification of transposable elements which activate gene expression in Pseudomonas cepacia. J Bacteriol. 1987 Jan;169(1):8–13. doi: 10.1128/jb.169.1.8-13.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair M. I., Holloway B. W. A chromosomally located transposon in Pseudomonas aeruginosa. J Bacteriol. 1982 Aug;151(2):569–579. doi: 10.1128/jb.151.2.569-579.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stanisich V. A., Bennett P. M., Richmond M. H. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J Bacteriol. 1977 Mar;129(3):1227–1233. doi: 10.1128/jb.129.3.1227-1233.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil M. L., Chamberlain C., Grant C. C. Molecular studies of Pseudomonas exotoxin A gene. Infect Immun. 1986 May;52(2):538–548. doi: 10.1128/iai.52.2.538-548.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Lee P. D., Kosuge T. Insertion sequence elements of Pseudomonas savastanoi: Nucleotide sequence and homology with Agrobacterium tumefaciens transfer DNA. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8263–8267. doi: 10.1073/pnas.83.21.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zabala J. C., García-Lobo J. M., Diaz-Aroca E., de la Cruz F., Ortiz J. M. Escherichia coli alpha-haemolysin synthesis and export genes are flanked by a direct repetition of IS91-like elements. Mol Gen Genet. 1984;197(1):90–97. doi: 10.1007/BF00327927. [DOI] [PubMed] [Google Scholar]