Abstract
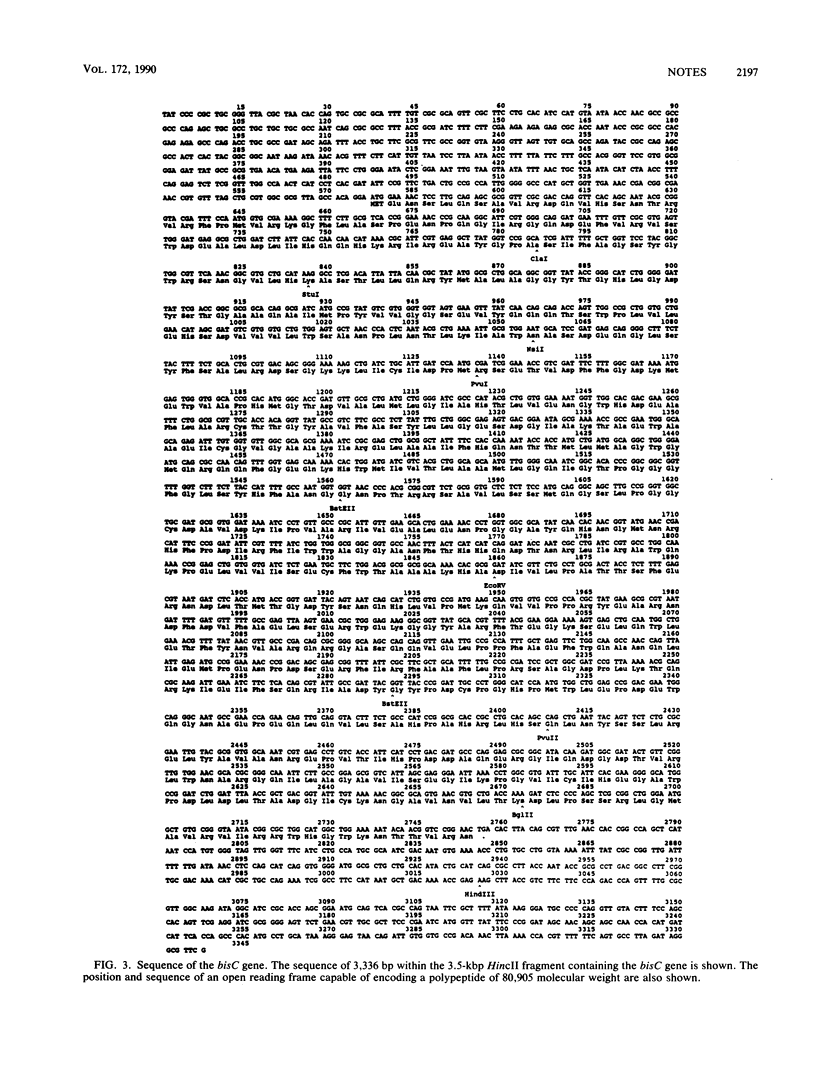
Clones of the Escherichia coli bisC locus have been isolated by complementing a bisC mutant for growth with d-biotin d-sulfoxide as a biotin source. The complementation properties of deletions and Tn5 insertions located the bisC gene to a 3.7-kilobase-pair (kbp) segment, 3.3 kbp of which has been sequenced. A single open reading frame of 2,178 bp, capable of encoding a polypeptide of molecular weight 80,905, was found. In vitro transcription of plasmids carrying the wild-type sequence and deletion and insertion mutants showed that BisC complementation correlated perfectly with production of a polypeptide whose measured molecular weight (79,000) does not differ significantly from 80,905.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bilous P. T., Cole S. T., Anderson W. F., Weiner J. H. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulphoxide reductase of Escherichia coli. Mol Microbiol. 1988 Nov;2(6):785–795. doi: 10.1111/j.1365-2958.1988.tb00090.x. [DOI] [PubMed] [Google Scholar]
- Bilous P. T., Weiner J. H. Dimethyl sulfoxide reductase activity by anaerobically grown Escherichia coli HB101. J Bacteriol. 1985 Jun;162(3):1151–1155. doi: 10.1128/jb.162.3.1151-1155.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brot N., Weissbach L., Werth J., Weissbach H. Enzymatic reduction of protein-bound methionine sulfoxide. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2155–2158. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A., Campillo-Campbell A. D., Barker D. Repression of biotin biosynthesis in Escherichia coli during growth on biotin vitamers. J Bacteriol. 1978 Jul;135(1):90–98. doi: 10.1128/jb.135.1.90-98.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casse F. Mapping of the gene chl-B controlling membran bound nitrate reductase and formic hydrogen-lyase activities in Escherichia coli K 12. Biochem Biophys Res Commun. 1970 May 11;39(3):429–436. doi: 10.1016/0006-291x(70)90596-6. [DOI] [PubMed] [Google Scholar]
- Claassen V. P., Oltmann L. F., Van't Riet J., Brinkman U. A., Stouthamer A. H. Purification of molybdenum cofactor and its fluorescent oxidation products. FEBS Lett. 1982 Jun 1;142(1):133–137. doi: 10.1016/0014-5793(82)80236-6. [DOI] [PubMed] [Google Scholar]
- Dubourdieu M., Andrade E., Puig J. Molybdenum and chlorate resistant mutants in Escherichia coli K12. Biochem Biophys Res Commun. 1976 Jun 7;70(3):766–773. doi: 10.1016/0006-291x(76)90658-6. [DOI] [PubMed] [Google Scholar]
- Ejiri S. I., Weissbach H., Brot N. The purification of methionine sulfoxide reductase from Escherichia coli. Anal Biochem. 1980 Mar 1;102(2):393–398. doi: 10.1016/0003-2697(80)90173-6. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Nealson K., Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983 Mar;32(3):773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Superoxide dismutases. Adv Enzymol Relat Areas Mol Biol. 1974;41(0):35–97. doi: 10.1002/9780470122860.ch2. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116(1):1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Haddock B. A., Mandrand-Berthelot M. A. Escherichia coli formate-to-nitrate respiratory chain: genetic analysis. Biochem Soc Trans. 1982 Dec;10(6):478–480. doi: 10.1042/bst0100478. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V. Characterization of the molybdenum cofactor of sulfite oxidase, xanthine, oxidase, and nitrate reductase. Identification of a pteridine as a structural component. J Biol Chem. 1980 Mar 10;255(5):1783–1786. [PubMed] [Google Scholar]
- Johnson M. E., Rajagopalan K. V. Involvement of chlA, E, M, and N loci in Escherichia coli molybdopterin biosynthesis. J Bacteriol. 1987 Jan;169(1):117–125. doi: 10.1128/jb.169.1.117-125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MELVILLE D. B. Biotin sulfoxide. J Biol Chem. 1954 Jun;208(2):495–501. [PubMed] [Google Scholar]
- Paluh J. L., Yanofsky C. High level production and rapid purification of the E. coli trp repressor. Nucleic Acids Res. 1986 Oct 24;14(20):7851–7860. doi: 10.1093/nar/14.20.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig J., Azoulay E. Etude génétique et biochimique des mutants résistant au Clo minus 3 (gènes chl A, chl B, chl C) C R Acad Sci Hebd Seances Acad Sci D. 1967 Apr 10;264(15):1916–1918. [PubMed] [Google Scholar]
- Puig J., Azoulay E., Pichinoty F. Etude génétique d'une mutation à effet pléiotrope chez l'Escherichia coli K 12. C R Acad Sci Hebd Seances Acad Sci D. 1967 Mar 13;264(11):1507–1509. [PubMed] [Google Scholar]
- Redfield R. J., Campbell A. M. Origin of cryptic lambda prophages in Escherichia coli K-12. Cold Spring Harb Symp Quant Biol. 1984;49:199–206. doi: 10.1101/sqb.1984.049.01.023. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. II. Mutations induced by bacteriophage lambda in Escherichia coli K12. J Mol Biol. 1973 Oct 25;80(2):297–314. doi: 10.1016/0022-2836(73)90174-5. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. Nucleotide sequence of the three major early promoters of bacteriophage T7. Nucleic Acids Res. 1979;6(5):1895–1907. doi: 10.1093/nar/6.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperl G. T., DeMoss J. A. chlD gene function in molybdate activation of nitrate reductase. J Bacteriol. 1975 Jun;122(3):1230–1238. doi: 10.1128/jb.122.3.1230-1238.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Tsuchiya T., Ishimoto M. Proton translocation coupled to trimethylamine N-oxide reduction in anaerobically grown Escherichia coli. J Bacteriol. 1981 Dec;148(3):762–768. doi: 10.1128/jb.148.3.762-768.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zubay G. In vitro synthesis of protein in microbial systems. Annu Rev Genet. 1973;7:267–287. doi: 10.1146/annurev.ge.07.120173.001411. [DOI] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]
- del Campillo-Campbell A., Campbell A. Molybdenum cofactor requirement for biotin sulfoxide reduction in Escherichia coli. J Bacteriol. 1982 Feb;149(2):469–478. doi: 10.1128/jb.149.2.469-478.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo-Campbell A., Dykhuizen D., Cleary P. P. Enzymic reduction of d-biotin d-sulfoxide to d-biotin. Methods Enzymol. 1979;62:379–385. doi: 10.1016/0076-6879(79)62244-9. [DOI] [PubMed] [Google Scholar]