Like athletic ability, metabolic tone is recognized to vary greatly among individuals. The ability to oxidize or store fat, build up muscle glycogen, withstand fasting, or respond to insulin with decreased blood glucose levels can indeed change greatly even within the same individual, based on such factors as age, frequency of exercise, and diet. Such changes in metabolic tone re flect the collective abundances and activities of hundreds of enzymes and other metabolic proteins, integrated within complex networks of metabolic pathways in multiple tissues. Much of this is driven by the overall profile of transcriptional activities of the genes encoding these proteins. Among the major nuclear regulators of metabolic gene expression, transcription factors that belong to the forkhead gene family have garnered increasing attention. The Drosophila embryo forkhead mutant displays dual projections at the head, hence the name, and this fly gene shares a 110-aa DNA binding domain with 39 forkhead genes in humans (for review, see ref. 1). At least six of these gene products (see ref. 2 for nomenclature), Foxo1, Foxo3a, Foxo4 (previously termed FKHR, FKHRL1, and AFX1) and Foxa1, Foxa2, and Foxa3 (previously termed HNF3α, HNF3β, and HNF3γ) appear to share major roles controlling the development of key metabolic tissues such as pancreatic islet cells and liver. They also regulate dozens of metabolic genes. For example, both Foxo and Foxa transcription factors are known to up-regulate key enzymes in the hepatic gluconeogenesis pathway, which prevents hypoglycemia in fasting (3, 4). In this issue of PNAS, Wolfrum et al. show how Foxa2a may be regulated by insulin (5).
Several lines of research have recently converged to highlight the Foxo subfamily as particularly exciting regulators of cell function. First, they are the closest orthologs to DAF-16, a Caenorhabditis elegans gene product found in genetic screens to be a target of insulin signaling through the phosphatidylinositol 3 (PI3)kinase pathway (6–8). DAF-16 was found to be a mediator of extended longevity in the worm, and this function is reversed by insulin receptor. In mammals, Foxo proteins were also shown to be critical regulators of cell survival and apoptosis as well as cell cycle progression at G1 (for reviews, see refs. 9 and 10). Additionally, Foxo1, Foxo3a, and Foxo4 contain three documented phosphorylation sites (one threonine and two serine) for the Akt (also denoted PKB) protein kinases downstream of the PI3-kinase pathway that is activated by insulin (11, 12). Phosphorylated Foxo proteins are localized in the cytoplasm, thus abrogating their transcriptional activity. Insulin can act through Akt to block Foxo-mediated transcription, attenuating expression of gene products that mediate gluconeogenesis or apoptosis, for example. In contrast, regulation of forkhead proteins in the Foxa subfamily by insulin has not been reported. Wolfrum et al. (5) show that Foxa2, but not Foxa1 and Foxa3, contains a single theonine site that is phosphorylated by Akt, and that this phosphorylation also causes cytoplasmic localization of Foxa2 in intact cells. Thus, a fourth forkhead protein is added to the list of insulin-regulated transcription factors.
Foxo proteins are critical regulators of cell survival, apoptosis, and cell cycle progression.
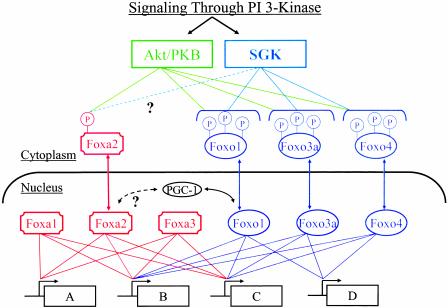
The data of Wolfrum et al. (5) add new dimensions and complexity to our understanding of insulin-regulated gene expression. The DNA-binding domains among forkhead members display a winged helix structure that reflects a helix–turn–helix core of three α-helices flanked by two loops or “wings” (13). Their sequences are highly similar or even identical in some cases, but significant variations can occur between subfamilies. Consistent with this profile, some genes display response elements that will bind multiple forkhead proteins, but other genes show selective sensitivity to one or another subfamily (14–16). This provides for redundancy in the regulation of some genes in a metabolic or developmental pathway, whereas others may be regulated by a unique forkhead protein. Fig. 1 depicts four genes (A, B, C, and D) in a hypothetical insulin-sensitive pathway in which genes B and C are regulated by both Foxa and Foxo proteins and gene A and gene D are selectively regulated by Foxa or Foxo family members, respectively. The discovery that Foxa2 is negatively regulated by insulin through Akt phosphorylation (5) extends the range of genes (i.e., gene A in Fig. 1) in such a pathway that can be controlled by the hormone. Importantly, in these studies, Foxa1 and Foxa3 were found not to be regulated by the insulin-sensitive Akt pathway. This finding leads to the prediction that constitutive regulation of certain insulin-sensitive genes by Foxa proteins is likely (genes A, B and C in Fig. 1). Thus, insulin's effects will be dampened by these influences of Foxa1 and Foxa3 in proportion to their expression levels within a tissue type and the extent to which Foxa-sensitive genes contribute to an overall pathway.
Fig. 1.
Regulation of the Foxa and Foxo transcription factors through PI3-kinase signaling. Protein kinases of the Akt/PKB and SGK families (for review, see ref. 35) downstream of 3′ polyphosphoinositides generated by PI3-kinase mediate phosphorylation and cytoplasmic localization of Foxa2 and three Foxo isoforms. Thus, insulin, IGF-1, and other regulators of PI3-kinase attenuate transcriptional activity of these proteins. Foxa2 regulates gene expression in the context of Foxa1 and Foxa3 actions, which are not regulated by PI3-kinase. Overlapping but distinct specificities of the Foxa and Foxo proteins for promoter sequences provide a complex network of transcriptional signals that can finely modulate sets of genes (A, B, C, and D) in a metabolic or developmental pathway.
The finding that Foxa2 is regulated by phosphorylation similar to the Foxo proteins also adds a potential level of complexity to the network of upstream signals emanating from the PI3-kinase pathway. As outlined in Fig. 1, the Akt/PKB and serum- and glucocorticoid-induced protein kinase (SGK) classes of protein kinases are activated through generation of 3′ polyphosphoinositides catalyzed by PI3-kinase. SGK expression is regulated by serum and glucocorticoids and controls epithelial sodium transport, apoptosis, and other processes. The protein kinase catalytic domains of SGK family members and their substrate specificities are somewhat similar to those of Akt/PKB. Both the Akt and SGK protein kinases have been shown to phosphorylate and negatively regulate the Foxo proteins (17). This provides redundancy as well as complementarity to the input signals for Foxo regulation, because these protein kinases show different preferences for specific phosphorylation sites on the Foxo proteins (10). Thus, the relative sensitivity of each of the three Foxo proteins to regulation by insulin is likely to vary depending on the relative expression levels of the Akt/PKB proteins versus SGK proteins. This will depend on cell type and conditions. Therefore, as the model in Fig. 1 implies, an important question raised by the results of Wolfrum et al. (5) is whether SGK protein kinases also phosphorylate Foxa2 and, if so, with what efficiency relative to Akt?
Adding further intrigue to this issue of protein kinase specificity for the forkhead proteins is the remarkable divergence in function between the Akt1 versus Akt2 isoforms. Akt1 functions in the determination of cell and organ size, based on the results of genetic manipulations in Drosophila (18) and on ectopic expression of Akt1 in beta cells of the mouse pancreas (19). Mice without Akt1 show normal glucose homeostasis but are small (20). In contrast, knockout of Akt2 in mice shows that it is required for normal glucose homeostasis but not for normal cell or organ size (21). These data are quite surprising based on the similar catalytic domains and apparent substrate specificities of these isoforms under cell-free conditions. Perhaps differential cellular localizations of these protein kinases or their substrates explains the divergent functions. Alternatively, phosphorylation site specificity on certain proteins may be more divergent between Akt1 and Akt2 than is expected, especially under conditions present in intact cells. If this is the case, might Foxa2 be such a substrate that shows differential sensitivity to Akt1 vs. Akt2, explaining in part the differences in actions between these protein kinases? It will be necessary to test this important question in future experiments.
It has recently been reported that Foxo1 acts to regulate the expression of enzymes in the gluconeogenesis pathway such as phosphoenolpyvurate carboxykinase (PEPCK) and glucose 6 phosphatase through direct interaction with the coactivator peroxisome proliferator-activated receptor γ coactivator (PGC)-1α (22). PCG-1 is itself highly up-regulated in the liver during fasting through the actions of hormones such as glucagon and glucocorticoids, and is a coactivator of the transcription factors cAMP response element binding (CREB) protein, liver-enriched transcription factor (HNF)-4α, and glucocorticoid receptor, which are required for full up-regulation of PEPCK (23–25). This coactivator also plays a key role in adaptive thermogenesis in brown fat and skeletal muscle (26). Remarkably, the association of Foxo1 with PGC-1 as well as the nuclear localization of Foxo1 are abrogated by Akt-mediated phosphorylation (22). Thus, insulin signaling through Akt can attenuate the effects of increased levels of PGC-1 in fasting and diabetic conditions through dissociation from Foxo1. It is not known whether Foxo3a and 4 are also regulated by PGC-1. These findings beg the question of whether PGC-1 is also a coactivator of Foxa2 (see Fig. 1). If so, might Akt also mediate dissociation of PGC-1 from Foxa2, thereby coordinating multiple pathways of PGC-1 actions? A recent additional twist to the connections between PGC-1 and Foxo proteins is the finding that PGC-1 promoter activity is itself regulated by Foxo1 in an insulin-dependent manner (27). Thus, insulin can apparently attenuate PGC-1 expression as well as its function through phosphorylation of Foxo proteins.
The above findings and conjectures reveal fertile experimental territory for future studies on Foxa2 function, especially in relation to its roles in the development and physiology of pancreatic islet cells, liver, and adipose tissue. Initial work indicating that Foxa2 functions in the development of the pancreas and liver (28, 29) was followed by results showing that tissue-specific deletion of Foxa2 in mouse pancreatic beta cells results in a profound hyperinsulinemic hypoglycemic condition (30). Two subunits of the beta cell ATP-sensitive potassium channel (KATP), frequently mutated genes linked to familial hyperinsulinism, were identified in these studies as novel targets of Foxa2. Thus, Foxa2 regulation by agents that regulate Akt, such as insulin and IGF-1, may have profound effects on beta cell insulin secretion and responsiveness to hypoglycemia. In the liver, Foxa2 may have a more prominent role in development than in maintaining adult tissue function, because liver-specific knockout of Foxa2 in mice has no effect on glucose homeostasis (31). However, this conclusion may require further evaluation because compensatory effects of other transcription factors could be at play, and Foxa3 certainly plays a role in modulating gluconeogenic enzymes (32) and GLUT2 (33) in liver. Finally, Foxa2 is expressed in preadipocytes and inhibits their differentiation into adipocytes (34). Remarkably, increased adiposity occurs when obesity is induced by diet in mice with haploinsufficiency in Foxa2 (Foxa2+/–), and defects in glucose and lipid metabolism are evident in these animals. These exciting new findings on the plethora of functions that Foxa proteins may play in metabolic tissues, combined with the discovery of Foxa2 regulation by Akt signaling, open new vistas for both understanding and intervening in metabolic diseases.
See companion article on page 11624.
References
- 1.Carlsson, P. & Mahlapuu, M. (2002) Dev. Biol. 250, 1–23. [DOI] [PubMed] [Google Scholar]
- 2.Kaestner, K. H., Knochel, W. & Martinez, D. E. (2000) Genes Dev. 14, 142–146. [PubMed] [Google Scholar]
- 3.Altomonte, J., Richter, A., Harbaran, S., Suriawinata, J., Nakae, J., Thung, S. N., Meseck, M., Accili, D. & Dong, H. (2003) Am. J. Physiol., in press. [DOI] [PubMed]
- 4.Kaestner, K. H. (2000) Trends Endocrinol. Metab. 11, 281–285. [DOI] [PubMed] [Google Scholar]
- 5.Wolfrum, C., Besser, D., Luca, E. & Stoffel, M. (2003) Proc. Natl. Acad. Sci. USA 100, 11624–11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin, K., Dorman, J. B., Rodan, A. & Kenyon, C. (1997) Science 278, 1319–1322. [DOI] [PubMed] [Google Scholar]
- 7.Ogg, S., Paradis, S., Gottlieb, S., Patterson, G. I., Tissenbaum, H. A. & Ruvkun, G. (1997) Nature 389, 994–999. [DOI] [PubMed] [Google Scholar]
- 8.Paradis, S. & Ruvkun, G. (1998) Genes Dev. 12, 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgering, B. M. T. & Kops, G. J. P. L. (2002) Trends Biochem. Sci. 27, 352–360. [DOI] [PubMed] [Google Scholar]
- 10.Tran, H., Brunet, A., Griffith, E. C. & Greenberg, M. E. (2003) Science STKE 2003, re5. [DOI] [PubMed] [Google Scholar]
- 11.Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., Anderson, M., Arden, K. C., Blenis, J. & Greenberg, M. E. (1999) Cell 96, 857–868. [DOI] [PubMed] [Google Scholar]
- 12.Takaishi, H., Konishi, H., Matsuzuki, Y., Ono, Y., Shirai, N., Saito, T., Kitamura, W., Ogawa, W., Kasuga, M., Kikkawa, U. & Nishizuka, Y. (1999) Proc. Natl. Acad. Sci. USA 96, 11836–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark, K. L., Halay, E. D., Lai, E. & Burley, S. K. (1993) Nature 364, 412–420. [DOI] [PubMed] [Google Scholar]
- 14.Overdier, D. G., Porcella, A. & Costa, R. H. (1994) Mol. Cell. Biol. 14, 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierrou, S., Hellqvist, M., Samuelsson, L., Enerback, S. & Carlsson, P. (1994) EMBO J. 13, 5002–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierrou, S., Enerback, S. & Carlsson, P. (1995) Anal. Biochem. 229, 99–105. [DOI] [PubMed] [Google Scholar]
- 17.Brunet, A., Park, J., Tran, L. S., Hemmings, B. A. & Greenberg, M. E. (2001) Mol. Cell. Biol. 21, 952–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verdu, J., Buratovich, M. A., Wilder, E. L. & Birnbaum, M. J. (1999) Nat. Cell Biol. 1, 500–506. [DOI] [PubMed] [Google Scholar]
- 19.Tuttle, R. L., Gill, N. S., Pugh, W., Lee, J. P., Koeberlein, B., Furth, E. E., Polonsky, K. S., Naji, A. & Birnbaum, M. J. (2001) Nat. Med. 7, 1133–1137. [DOI] [PubMed] [Google Scholar]
- 20.Cho, H., Thorvaldsen, J. L., Chu, Q., Feng, F. & Birnbaum, M. J. (2001) J. Biol. Chem. 276, 38349–38352. [DOI] [PubMed] [Google Scholar]
- 21.Cho, H., Mu J., Kim, J. K., Thorvaldsen, J. L., Chu, Q., Crenshaw, E. B., III, Kaestner, K. H., Bartolomei, M. S., Shulman, G. I. & Birnbaum, M. J. (2001) Science 292, 728–1731. [DOI] [PubMed] [Google Scholar]
- 22.Puigserver, P., Rhee, J., Donovan, J., Walkey, C. J., Yoon, J. C., Oriente, F., Kitamura, Y., Altomonte, J., Dong, H., Accili, D. & Spiegelman, B. M. (2003) Nature 423, 550–555. [DOI] [PubMed] [Google Scholar]
- 23.Rhee, J., Inoue, Y., Yoon, J. C., Puigserver, P., Fan, M., Gonzalez, F. J. & Spiegelman, B. M. (2003) Proc. Natl. Acad. Sci. USA, 100, 4012–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herzig, S., Long, F., Jhala, U. S., Hedrick, S., Quinn, R., Bauer A., Rudolph, D., Schutz, G., Yoon C., Puigserver, P., et al. (2001) Nature 413, 179–183. [DOI] [PubMed] [Google Scholar]
- 25.Yoon, J. C., Puigserver, P., Chen, G., Donovan, J., Wu, Z., Rhee, J., Adelmant, G., Stafford, J., Kahn, C. R., Granner, D. K., et al (2001) Nature 413, 131–138. [DOI] [PubMed] [Google Scholar]
- 26.Puigserver, P. & Spiegelman, B. M. (2003) Endocr. Rev. 24, 78–90. [DOI] [PubMed] [Google Scholar]
- 27.Daitoku, H., Yamagata, K., Matsuzaki, H., Hatta, M. & Fukamizu, A. (2003) Diabetes 52, 642–649. [DOI] [PubMed] [Google Scholar]
- 28.Ang, S. L. & Rossant, J. (1994) Cell 78, 561–574. [DOI] [PubMed] [Google Scholar]
- 29.Duncan, S. A., Navas, M. A., Dufort, D., Rossant, J. & Stoffel, M. (1998) Science 281, 692–695. [DOI] [PubMed] [Google Scholar]
- 30.Sund, N. J., Vatamaniuk, M. Z., Casey, M., Ang, S. L., Magnuson, M. A., Stoffers, D. A., Matschinsky, F. M. & Kaestner, K. H. (2001) Genes Dev. 15, 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sund, N. J., Ang, S. L., Sackett, S. D., Shen, W., Daigle, N., Magnuson, M. A. & Kaestner, K. H. (2000) Mol. Cell. Biol. 20, 5175–5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaestner, K. H., Hiemisch, H. & Schutz, G. (1998) Mol. Cell. Biol. 18, 4245–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen, W., Scearce, M., Brestelli, J. E., Sund, N. J. & Kaestner, K. H. (2001) J. Biol. Chem. 276, 42812–42817. [DOI] [PubMed] [Google Scholar]
- 34.Wolfrum, C., Shih, D. Q., Kuwajima, S., Norris, A. W., Kahn, C. R. & Stoffel, M. (2003) J. Clin. Invest. 112, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang, F. & Cohen, P. (2001) Science STKE 2001, re 17. [DOI] [PubMed] [Google Scholar]