Abstract
Conjugative plasmid transfer is an important mechanism for diversifying prokaryotic genomes and disseminating antibiotic resistance. Relaxases are conjugative plasmid-encoded proteins essential for plasmid transfer. Relaxases bind and cleave one plasmid strand site- and sequence-specifically before transfer of the cleaved strand. TraI36, a domain of F plasmid TraI that contains relaxase activity, binds a plasmid sequence in single-stranded form with subnanomolar KD and high sequence specificity. Despite 91% amino acid sequence identity, TraI36 domains from plasmids F and R100 discriminate between binding sites. The binding sites differ by 2 of 11 bases, but both proteins bind their cognate site with three orders of magnitude higher affinity than the other site. To identify specificity determinants, we generated variants having R100 amino acids in the F TraI36 background. Although most retain F specificity, the Q193R/R201Q variant binds the R100 site with 10-fold greater affinity than the F site. The reverse switch (R193Q/Q201R) in R100 TraI36 confers a wild-type F specificity on the variant. Nonadditivity of individual amino acid and base contributions to recognition suggests that the specificity difference derives from multiple interactions. The F TraI36 crystal structure shows positions 193 and 201 form opposite sides of a pocket within the binding cleft, suggesting binding involves knob-into-hole interactions. Specificity is presumably modulated by altering the composition of the pocket. Our results demonstrate that F-like relaxases can switch between highly sequence-specific recognition of different sequences with minimal amino acid substitution.
Conjugative plasmids are important conduits for genetic transfer between prokaryotes. During bacterial conjugation, a conjugative plasmid directs transfer of a copy of itself, in single-stranded form, to a recipient cell (reviewed in refs. 1–3). Despite the sequence diversity of conjugative plasmids that have been studied, many elements of the transfer process are conserved (3), including the involvement of relaxases. Relaxases, also called mobilization proteins or nickases, are essential for conjugation (4–6). These proteins bind and cleave one DNA strand at the plasmid origin of transfer (oriT). The cleaved strand is transferred to the recipient. Relaxases act as part of the relaxosome, a complex of multiple proteins and plasmid DNA (6–8). The relaxosome may serve to generate the single-stranded DNA (ssDNA) substrate that the relaxase targets. Relaxases cleave ssDNA in a Mg2+-dependent transesterification reaction that proceeds through a stable phosphotyrosyl intermediate (9–15). At the end of conjugative transfer, relaxases are thought to ligate the plasmid ends together (9, 16–19).
Relaxases exhibit sequence specificity for their cognate oriT sequences (9, 11, 12, 20–26). We previously demonstrated, by using the 36-kDa relaxase domain of F TraI (TraI36), an N-terminal 330-aa fragment of F factor TraI (27), that this specificity can be remarkable. F TraI36 binds a single-stranded F oriT sequence with a subnanomolar KD, and some single-base substitutions over a 10-base region can reduce affinity by between 10- and 10,000-fold (24). In vivo, this specificity is reflected in the poor efficiency with which F TraI mobilizes plasmids containing the TraI-binding site for the highly homologous R100 plasmid, despite a difference of only two bases (24, 28). To better understand the basis of ssDNA recognition by relaxases, we set out to identify F TraI36 amino acids that contribute to ssDNA recognition. Here we demonstrate that despite the 91% amino acid sequence identity shared by F factor and R100 TraI36, the two proteins have easily detected differences in specificity. We then describe the results from a panel of TraI36 variants that identify specificity determinants. We found that just two amino acid residue differences account for the two base difference in ssDNA binding and cleavage specificity.
Materials and Methods
Protein Engineering and Purification. Primer sequences are available on request. All cloned gene segments and mutations were confirmed by DNA sequencing. The F TraI36 expression construct (pET24a-traI36) was engineered previously (27). For R100 TraI36 expression, the N-terminal 331-codon region of R100 traI was PCR amplified. Primers encoded an NdeI site overlapping the start codon and an EcoRI site after a stop codon engineered at codon 332. The NdeI/EcoRI-digested PCR product was ligated into NdeI/EcoRI-digested pNEB193 (New England Biolabs), transformed into the XL10 Gold (Stratagene) E. coli strain, and plasmids containing inserts were identified by α complementation. The R100 traI36 gene was excised from pNEB193, ligated into NdeI/EcoRI-digested pET24a(+) (Novagen), and transformed into strain XL10 Gold. The 3′-most codon was removed by PCR as described (27).
Gene fragments for chimera R100/F/F were generated by NdeI/PstI digestion of the TraI36 expression vectors, purified by electrophoresis through low melt agarose, isolated by using the Wizard PCR Preps DNA Purification System kit (Promega), and ligated. The R100 insert fragment for chimera F/F/R100 was PCR-amplified by using R100 traI36 as template, and cloned into purified StuI/EcoRI-digested pET24a-traI36. Constructs were transformed into strain TB1. An unintended M278T mutation in chimera F/F/R100 was corrected by using the QuikChange site-directed mutagenesis kit (Stratagene).
Single, double, and triple mutations were generated by using the QuikChange kit. An “F” or “R” preceding the variant description indicates F or R100 background, respectively. The F-Q193R/R201Q variant was PCR-generated by using the F-Q193R traI36 variant as the template. Variants F-E153D/Q193R/R201Q and F-I185S/Q193R/R201Q were PCR-generated from F-Q193R/R201Q, R-R193Q/Q201R from R-Q201R, and R-D153E/R193Q/Q201R from R-R193Q/Q201R.
Expression plasmids were transformed into strain BL21(DE3). Proteins were expressed and purified as described for wild type F TraI36 (27), and concentrated by using CentriCon10 or Centricon Plus-20 filters (Amicon).
Oligonucleotide Synthesis and Purification. Sequences of oligonucleotides used in binding and cleavage assays are shown in Fig. 1. Oligonucleotides were purchased from Integrated DNA Technologies, Inc. 3′ carboxytetramethylrhodamine (TAMRA) labels were incorporated by using TAMRA-CPG columns. Labeled oligonucleotides and some PCR primers were purified by polyacrylamide gel electrophoresis as described (29). 32P 5′-end labeling was performed as described (29).
Fig. 1.
Sequences of the binding site oligonucleotides. The 5′ end of the F sequence starts at 154′, where the numbering is according to Frost et al. (2). The prime indicates the ssDNA sequence bound by TraI is on the “bottom” strand versus the numbered “top” strand. Positions 147′ and 145′, which differ between F and R100, are in bold, and bases differing from the F sequence are underscored. The vertical line indicates nic, the site of DNA cleavage by TraI.
DNA Binding and Cleavage. Affinity measurements were performed by titrating protein into 4 nM solutions of fluorophore-labeled oligonucleotide and following changes in fluorescence anisotropy and emission intensity as described (24, 30), or, for highest affinity interactions, in a buffer with 250 mM NaCl replacing the 100 mM NaCl normally used. Data were fit by using the global fitting algorithm spectrabind (31, 32) as described (30). Cleavage of single-stranded, 22-base oligonucleotide with F factor or R100 sequence by various TraI36 proteins was examined by using a nicking assay (18) as described (24). Despite repeated purification attempts, some variants copurified with a small amount of a nonspecific nuclease activity. Binding and cleavage assays were repeated in the presence of 1 μM nonspecific oligonucleotide for these variants and the wild-type F TraI36 with no differences in results.
CD. CD experiments were performed by using 2.5 or 5 μM protein in 20 mM sodium phosphate (pH 7.5), 100 mM NaCl, and 0.1 mM EDTA on an AVIV215 CD Spectrometer. Wavelength scans were performed at 25°C with 1-nm steps and 1-s signal averaging at each wavelength. Thermal denaturations were observed at 222 nm by using 1°C steps from 25°Cto75°C, 60-s temperature equilibrations, and 30-s signal averaging.
Results and Discussion
Wild-Type F and R100 TraI36 Binding. In vivo plasmid nicking and transfer experiments have demonstrated that TraI proteins from F and R100 can distinguish the two-base difference between their cognate TraI binding site (Fig. 1) and the site from the other plasmid (20, 24, 26, 28). To quantify the binding differences, an expression construct for the R100 TraI relaxase domain (TraI36) was generated, the protein was purified, and its ssDNA binding was compared with that of the previously generated F TraI36 (27). The TraI36 domains consist of the N-terminal 330 TraI amino acids, and share 91% amino acid sequence identity.
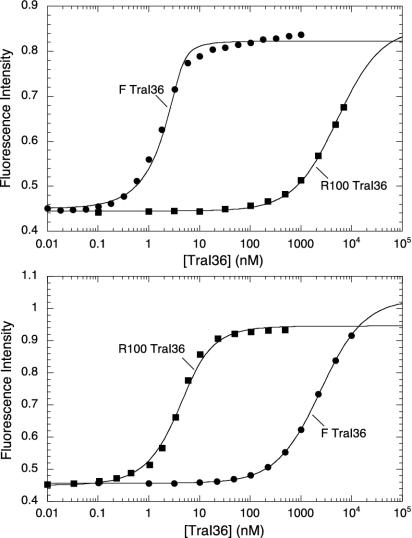
Dissociation constants for the interactions between F and R100 TraI36 and the TraI-binding sites from F and R100 oriT regions were measured by following anisotropy and total emission intensity changes of TAMRA-labeled single-stranded oligonucleotides on titration of protein. Because the cleavage activity requires Mg2+ and the binding assays are performed in absence of Mg2+, the binding is reversible and the assay measures an equilibrium binding constant. Dissociation constants are listed in Table 1, and representative binding curves are shown in Fig. 2. F TraI36 binds to the F oriT with a 0.55 nM KD, in agreement with previous measurements (24, 30). F TraI36 binds the R100 oriT oligonucleotide with an estimated 3.3 μM KD, correlating well with previous competition studies (24). R100 TraI36 is also specific for its cognate site, binding the R100 oriT oligonucleotide with a 2.4 nM KD and the F oriT oligonucleotide with an estimated 3.2 μM KD. (In experiments yielding KD values >1 μM, binding frequently did not reach saturation. However, because both fluorescence intensity and anisotropy data were simultaneously fit, we can estimate KD values that fall between 1 and 5 μM. KD values above this range are listed as >5 μM.)
Table 1. Binding constants of wild-type and chimeric TraI36 proteins.
| F oriT
|
R100 oriT
|
|||
|---|---|---|---|---|
| TraI36 protein | KD (nM) ± SD | N | KD (nM) ± SD | N |
| F factor | 0.55 ± 0.07 | 2 | 3,300 ± 650 | 2 |
| R100 | 3,200 ± 1,400 | 2 | 2.4 ± 0.15 | 2 |
| R100/F/F* | 6.3 ± 0.2 | 2 | >5,000 | 1 |
| F/F/R100† | 0.52 ± 0.3 | 2 | >5,000 | 1 |
Chimera R100/F/F contains amino acids 1-44 from R100 TraI36.
Chimera F/F/R100 contains amino acids 232-330 from R100 TraI36.
Fig. 2.
Differing specificities of the wild-type F and R100 TraI36 proteins. Emission intensity of 3′ 22-base TAMRA-labeled binding site oligonucleotides increased on binding wild-type F (circles) and R100 (squares) TraI36. Oligonucleotides include TraI binding sites from F (Upper) and R100 (Lower) oriT. Intensity data were adjusted for fluorophore dilution and normalized for initial intensity (arbitrary units) relative to each other. Solid lines indicate results of the simultaneous fit of intensity and anisotropy (not shown) data to a 1:1 binding model by using spectrabind (31, 32).
Binding of Chimeras and F TraI36 Variants. To identify specificity determinants for the proteins, initially two chimeric molecules were generated. Chimera R100/F/F contained R100 sequence for the N-terminal 44 aa, including 6 amino acid differences between the F and R100 proteins. The affinity of this protein for F oriT is reduced 10-fold but its affinity for R100 oriT is unchanged, relative to wild-type F TraI36 (Table 1). Chimera F/F/R100 contained R100 sequence for the C-terminal 98 aa, including 9 amino acid differences. This chimera has wild-type F affinity and specificity (Table 1).
The results from the chimeric proteins suggested that the major specificity determinants are located between amino acids 44 and 232. F and R100 TraI36 differ at 14 positions in this region. Variants having one to three R100 amino acids in the F background (designated by an F preceding the amino acid substitution) were generated and examined. To help speed identification of specificity determinants, multiple substitutions were combined in a single variant when the amino acid positions were close enough to make it experimentally feasible. The KD values for the variants are shown in Table 2. Variants F-A160V, F-R197K/L198I, F-E202S, F-K203E, and F-K205R/E206Q/Q207R retained F specificity, with KD values for both binding sites within 2-fold of the values for wild-type F protein.
Table 2. Binding constants of F TraI36 variants.
| F oriT site
|
R100 oriT site
|
|||
|---|---|---|---|---|
| TraI36 protein | KD (nM) ± SD | N | KD (nM) ± SD | N |
| F factor | 0.55 ± 0.067 | 2 | 3,300 ± 650 | 2 |
| R100 | 3,200 ± 1400 | 2 | 2.4 ± 0.15 | 2 |
| F-R75K | <0.5 | 3 | 1,600 ± 660 | 2 |
| F-E153D | <0.5 | 3 | 390 ± 3.9 | 2 |
| F-A160V | 1.0 ± 0.22 | 2 | 4,000 | 1 |
| F-I185S | 1.2 ± 0.19 | 2 | 200 ± 0.71 | 2 |
| F-Y190L | 19 ± 0.60 | 2 | 4,400 | 1 |
| F-Q193R | 860 ± 30 | 2 | 3,600 ± 490 | 2 |
| F-R198K/L199I | 0.42 ± 0.021 | 2 | 1,600 | 1 |
| F-R201Q | 480 ± 37 | 2 | >5,000 | 2 |
| F-E202S | 0.49 ± 0.27 | 2 | >5,000 | 1 |
| F-K203E | 0.99 ± 0.25 | 2 | >5,000 | 1 |
| F-K205R/E206Q/Q207R | 0.33 ± 0.13 | 2 | >5,000 | 1 |
| F-Q193R/R201Q | >5,000 | 2 | 460 ± 6.4 | 2 |
| F-I185S/Q193R/R201Q | >5,000 | 2 | 160 ± 8.5 | 2 |
| F-E153D/Q193R/R201Q | 1,500 ± 39 | 2 | 72 ± 7.5 | 2 |
| F-E153D/I185S/Q193R/R201Q | >5,000 | 1 | 120 ± 11 | 2 |
Variants F-E153D and F-R75K show a general increase in affinity. F-E153D binds the R100 oriT oligonucleotide with 8-fold higher affinity, and F-R75K with 3-fold higher affinity, than wild-type F TraI36. The variants also bind the F oriT oligonucleotide with increased affinity. Assay conditions, specifically the 4 nM concentration of oligonucletide required for sufficient fluorescent signal, make it difficult to measure affinities significantly higher than that of wild-type F. Therefore, relative affinities of F, F-E153D, and F-R75K TraI36 for F oriT were determined by using higher salt concentrations (250 mM vs. 100 mM NaCl), at which F TraI36 has reduced affinity for ssDNA (J. C. Stern and J.F.S., unpublished data). These results (not shown) confirm that the two variants have a severalfold increased affinity for F oriT, indicating that these variants have increased affinity but essentially unaltered specificity.
Four single amino acid substitutions cause distinct changes in binding specificity. F-I185S shows a 2-fold reduction in affinity for F oriT but a 16-fold increased affinity for the R100 oriT sequence, relative to F TraI36. F-Y190L and F-Q193R have 35-fold and 1,500-fold decreased affinity, respectively, for the F oriT oligonucleotide, but unchanged affinity for the R100 oriT, relative to F TraI36. F-R201Q has an 870-fold reduction in affinity for F oriT. The F-R201Q variant also appears to have reduced affinity for R100, although the affinity is low and difficult to accurately quantify.
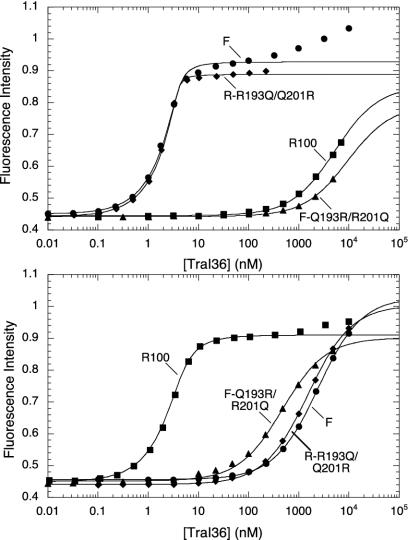
Because the substitutions at 193 and 201 cause the most significant changes in binding, we combined them in the F-Q193R/R201Q variant. This protein (Table 2 and Fig. 3) has a switched preference of binding sites, with at least 10-fold better binding of the R100 site (KD = 460 nM) than the F site (>5 μM). These two amino acid substitutions are sufficient to change the specificity from F to an “R100-like” specificity, although the affinity of the variant for the R100 site is still considerably reduced relative to that of the R100 TraI36 protein. Clearly, the altered specificity results from a combinatorial effect of the two substitutions.
Fig. 3.
Variant TraI36 proteins demonstrate altered specificity. Emission intensity of a 3′ 22-base TAMRA-labeled binding site oligonucleotides was followed during titration of wild-type F (circles), R100 (squares), F-Q193R/R201Q (triangles), or R-R193Q/Q201R (diamonds) TraI36 proteins. Oligonucleotides contained F (Upper) or R100 (Lower) oriT sequences. Intensity data were adjusted for fluorophore dilution and normalized for initial intensity (arbitrary units) relative to each other. Solid lines indicate results of the simultaneous fit of intensity and anisotropy (not shown) data to a 1:1 binding model by using spectrabind (31, 32).
Variants with three and four amino acid substitutions were engineered and tested to determine the effect of altering amino acids within the context of the Q193R/R201Q substitutions. F-E153D/Q193R/R201Q has a 6-fold, and F-I185S/Q193R/R201Q a 3-fold, enhanced affinity for the R100 site, relative to the F-Q193R/R201Q variant (Table 2). F-E153D/Q193R/R201Q also has improved binding to the F site, relative to the double variant, whereas F-I185S/Q193R/R201Q binding to F oriT is too low to accurately measure. Combining both the I185S and E153D substitutions with Q193R/R201Q fails to improve the binding to the R100 oriT sequence. Consistent with the results from the F-Y190L variant, F-Y190L/Q193R/R201Q has poor binding to both the F and R100 oriT sequences.
The Q193R/R201Q switch is the key to changing the ssDNA-binding specificity from F to R100-like. Adding other amino acid substitutions improve binding, but the F TraI36 variants still demonstrate only a 20-fold higher affinity for the R100 site relative to the F site compared with the 1,400-fold difference exhibited by R100 TraI36. Clearly, additional substitutions are required to fully convert the F TraI36 specificity to that of R100 TraI36. Some of these substitutions may, like I185S, exert different effects in different contexts, making them more difficult to identify through individual amino acid changes.
Binding of R100 Variants. The results for F TraI36 indicate that amino acids at 193 and 201 are essential to the specificity difference between F and R100. Given this, placing the residues from F into the R100 background might shift the specificity of the protein from R100 to F. Results for a series of variants having F amino acids substituted into the R100 background, which are designated by an “R” preceding the amino acid substitutions, are listed in Table 3.
Table 3. Binding constants of R100 TraI36 variants.
| F oriT
|
R100 oriT
|
|||
|---|---|---|---|---|
| TraI36 protein | KD (nM) ± SD | N | KD (nM) ± SD | N |
| F factor | 0.55 ± 0.067 | 2 | 3,300 ± 650 | 2 |
| R100 | 3,200 ± 1,400 | 2 | 2.4 ± 0.15 | 2 |
| R-R193Q | 46 ± 0.3 | 2 | 350 ± 20 | 2 |
| R-Q201R | 120 ± 29 | 2 | 600 ± 5.7 | 2 |
| R-R193Q/Q201R | <0.5 | 2 | 1,750 ± 230 | 2 |
| R-D153E/R193Q/Q201R | <0.5 | 2 | >5,000 | 2 |
Variants R-R193Q and R-Q201R both bind F oriT with at least 5-fold higher affinity than R100 oriT. Each of these single substitutions reduce binding to the R100 oriT site by at least 150-fold and increase binding to the F oriT site by at least 25-fold, relative to wild-type R100. The variants R-R193Q/Q201R (Fig. 3) and R-D153E/R193Q/Q201R both display essentially F specificity and affinity, each binding at least 3,000-fold better to the F sequence than the R100 sequence. The high affinities of these R100 variants for the F sequence were confirmed by measuring binding at high salt conditions (data not shown). The conversion from the R100 to the F specificity is therefore more easily accomplished than the conversion from F to R100.
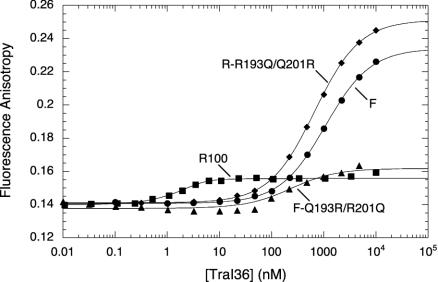
Anisotropy Differences. In addition to displaying altered binding, variants F-Q193R/R201Q and R-R193Q/Q201R also cause different effects on fluorescence anisotropy on binding the TAMRA-labeled oligonucleotides. As shown in Fig. 4 for the R100 oriT oligonucleotide, binding by R100 TraI36 causes a relatively small increase (from 0.14 to 0.15) in fluorophore anisotropy. In contrast, binding by F TraI36 causes a considerably larger increase in fluorescence anisotropy (from 0.14 to 0.23). Variant R-R193Q/Q201R, which has the F-binding specificity, also causes the F-like large increase in fluorescence anisotropy on binding. The F-Q193R/R201Q variant, which preferentially binds the R100 site, shows an R100-like small relative increase in anisotropy. A similar effect is seen with the F oriT oligonucleotide, with F TraI36 and R-R193Q/Q201R causing small increases in fluorescence anisotropy of the oligonucleotide and R100 TraI36 and F-Q193R/R201Q causing larger increases (not shown). This phenomenon appears to be related to specificity rather than absolute affinity because F-Q193R/R201Q and R-R193Q/Q201R bind R100 oriT with only a 4-fold difference in affinity but cause different effects on anisotropy.
Fig. 4.
Differences in fluorescence anisotropy changes correspond to different specificities. Shown are changes in fluorescence anisotropy of 3′ 22-base TAMRA-labeled R100 oriT-binding site oligonucleotide on binding of wild-type F (circles), R100 (squares), F-Q193R/R201Q (triangles), or R-R193Q/Q201R (diamonds) TraI36 proteins. Variants F-Q193R/R201Q and R-R193Q/Q201R appear to confer the converse swap in anisotropic properties on binding. Solid lines indicate results of the simultaneous fit of intensity and anisotropy (not shown) data to a 1:1 binding model by using spectrabind (31, 32).
These results indicate that, in this system, anisotropic differences not only distinguish between bound and unbound states, but also report on a difference between the physical properties of the protein/ssDNA complexes. The basis of this phenomenon is unclear. There may be differences in the conformations of the bound oligonucleotides that could affect local motion of the fluorophore and, thus, anisotropy. There may also be different interactions between the fluorophore and the DNA and/or protein in the different complexes, altering anisotropic behavior by altering the motion or fluorescent lifetime of the fluorophore. Analogous results were obtained for the F TraI36 variant F-R150A, which causes an increase in fluorescence intensity but not steady-state anisotropy on binding the 22-base 3′-TAMRA labeled oriT oligonucleotide (30). It is also possible that there is an oligomerization of the protein induced by less specific interactions, although we have no other evidence consistent with this model.
Fine Specificity of ssDNA Recognition. Binding of the variants to oligonucleotides having single base substitutions was examined (Table 4). The base numbering is for the F sequence according to Frost and colleagues (2), with a prime indicating the “bottom” (cleaved) DNA strand.
Table 4. Binding constants of F and R100 TraI36 variants for F and R100 oriT sites.
| Amino acid at
|
||||||
|---|---|---|---|---|---|---|
| TraI36 protein | 193 | 201 | 145′G/147′G (F oriT), KD ± SD* | 145′T/147′G, KD ± SD | 145′G/147′A, KD ± SD | 145′T/147′A (R100 oriT), KD ± SD |
| F factor | Q | R | 0.55 ± 0.067 | 2,500 ± 330 | 120 ± 6 | 3,300 ± 650 |
| R-R193Q/Q201R | Q | R | <0.5 | 620 ± 80 | 11 ± 5 | 1,750 ± 230 |
| R-R193Q | Q | Q | 46 ± 0.3 | 3.4 ± 0.4 | >5,000 | 350 ± 20 |
| R-Q201R | R | R | 120 ± 29 | 1,400 ± 540 | 32 ± 14 | 600 ± 5.7 |
| F-E153D/Q193R/R201Q | R | Q | 1,500 ± 39 | 170 ± 28 | >5,000 | 72 ± 7.5 |
| R100 | R | Q | 3,200 ± 1,400 | 2.8 ± 1.1 | 3,900 ± 440 | 2.4 ± 0.15 |
Boldface indicates bases that differ from the F oriT sequence.
Values an average of at least two measurements.
The strongest correlation observed was between the amino acid at position 201 and the base at position 145′, suggesting a direct interaction between the two. Those proteins with R201 (F, R-Q201R, R-R193Q/Q201R) bind sequences with 145′G at least 10-fold better than otherwise identical sequences with 145′T. Those proteins with Q201 (R100; R-R193Q; F-E153D/Q193R/R201Q) bind oligonucleotides with 145′T at least 9-fold better than otherwise identical oligonucletides with 145′G. Consistent with these findings, DNA-binding proteins often contact G bases through Arg side chains, and T bases are more frequently contacted by Gln than Arg side chains (33).
Other correlations between amino acids and specificity are relatively weak. Variant R-R193Q, which has Gln at both 193 and 201, exhibits a 100-fold preference in binding to sequences with 147′G versus 147′A. In contrast, those proteins with Q193 and R201 (F and R-R193Q/Q201R) bind with 20-fold higher affinity to the F oriT (145′G/147′G) over the 145′G/147′A sequence, but show no significant preference for a 147′G over a 147′A when in the context of 145′T. Those proteins having R193 and Q201 do not distinguish significantly between 147′G and 147′A. R-Q201R, which has Arg at both 193 and 201, shows a small (2- to 4-fold) preference for 147′A over 147′G.
These data indicate that F TraI36 has more specific recognition than R100. R100 binds its cognate oriT site with >1,000-fold higher affinity than the F site, yet this specificity is due largely to the identity of the base at position 145′. In contrast, F TraI36 discriminates between the F and R100 oriT bases at both 145′ and 147′.
The sum of the data suggest that the specificity is influenced by the amino acids at 193 and 201, and can also be influenced by the bases at 145′ and 147′. This suggests that these bases and amino acids interact, directly or indirectly, to determine specificity.
CD Studies of Variants. The stability of the proteins was assessed to examine the possibility that the altered recognition was related to altered stability. CD wavelength spectra for wild-type F and R100 TraI36, and variants F-Q193R/R201Q, F-E153D/Q193R/R201Q, and R-R193Q/Q201R are similar (not shown). These proteins also have similar transitions in thermal denaturation experiments (not shown), indicating similar stabilities and suggesting that the altered specificities are not explained by altered protein stabilities. Both F (27) and R100 (not shown) TraI36 show considerable hysteresis during thermal and chemical denaturation and renaturation, precluding calculation of the Gibbs free energy of unfolding.
Oligonucleotide Nicking. The correlation between oligonucleotide binding and nicking was assessed because F and R100 TraI36 exhibit different levels of specificity in oligonucleotide cleavage, making cleavage a further measure of the specificity of these enzymes. F TraI36 cleaves its cognate oriT sequence well, but cleaves the R100 sequence poorly (Figs. 5 and 6). R100 TraI36 cleaves its cognate sequence preferentially, but can also cleave the F sequence relatively well. The ssDNA cleavage specificity results for F and R100 TraI36 are consistent with the results of in vivo nicking and transfer experiments using the full-length proteins. R100 TraI nicks and efficiently transfers chimeric plasmids that include the F TraI binding site (28), whereas F TraI is highly specific in its ssDNA cleavage and plasmid mobilization activities, and interacts less effectively with the R100 sequence (24, 28).
Fig. 5.
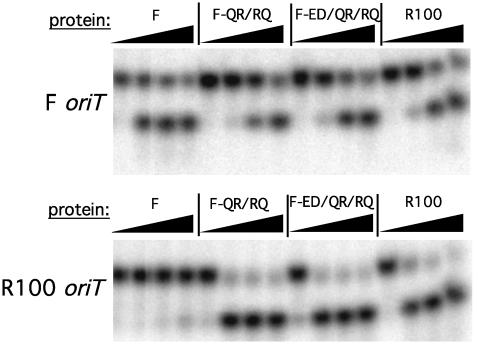
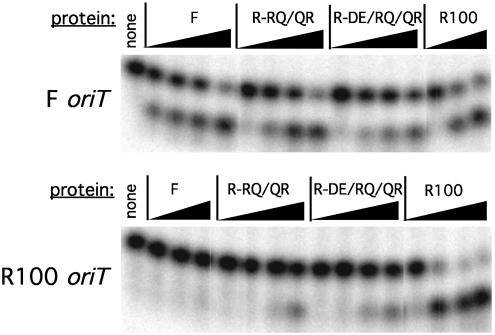
Oligonucleotide cleavage specificity of variant F-Q193R/R201Q correlates with binding specificity. A 5′ radiolabeled oligonucleotide was incubated with protein in a Mg2+-containing buffer, the reaction was stopped with SDS, and the products were electrophoresed through a denaturing gel. Cleavage is indicated by conversion of the oligonucleotide from a 22-base to a 14-base fragment. Shown are F (Upper) and R100 (Lower) oriT oligonucleotides cleaved by wild-type F, F-Q193R/R201Q (F-QR/RQ), F-E153D/Q193R/R201Q (F-ED/EQ/RQ), and wild-type R100 in the presence of 1 μM unlabeled, nonspecific DNA. Protein concentrations are 10 nM, 100 nM, 1 μM, and 10 μM.
Fig. 6.
Oligonucleotide cleavage specificity of variant R-R193Q/Q201R correlates with binding specificity. Shown are F (Upper) and R100 (Lower) oriT oligonucleotides cleaved by wild-type F, R-R193Q/Q201R (R-RQ/QR), R-D153E/R193Q/Q201R (R-DE/RQ/QR), and wild-type R100 TraI36. The left lane is the oligonucleotide in the absence of protein. The protein concentrations are 10 nM, 100 nM, 1 μM, and 10 μM, except for R100 TraI36 in Upper and F TraI36 in Lower, which do not contain a 10 nM sample.
All variants tested retain overall specificity for the TraI binding site as demonstrated by the presence of a single cleavage product in each assay, with the cleavage products having equivalent electrophoretic mobilities (Figs. 5 and 6). Variants F-Q193R/R201Q and F-E153D/Q193R/R201Q have reduced apparent cleavage activity on the F oriT oligonucleotide, relative to the F parent, to a level near that of R100 TraI36 (Fig. 5 Upper). These variants also show greatly increased apparent cleavage activity on the R100 oriT oligonucleotide, to the level of R100 TraI36 (Fig. 5 Lower). The F single variants show reduced apparent cleavage activity on the F oriT oligonucleotide and slight (F-R201Q) or no (F-Q193R) appreciable gain of apparent cleavage activity on the R100 oriT oligonucleotide (data not shown).
R100 background variants R-R193Q/Q201R and R-D153E/ R193Q/Q201R display cleavage characteristics intermediate between F TraI36 and their R100 TraI36 parent. Both variants generate more F oriT cleavage product at 100 nM protein than R100 TraI36, although the difference is slight for variant R-R193Q/Q201R (Fig. 6 Upper). Both variants have decreased apparent activity against the R100 oriT oligonucleotide, relative to R100 TraI36, but still had more apparent activity than F TraI36 (Fig. 6 Lower). R-R193Q and R-Q201R display greater apparent cleavage activity on F oriT than wild-type R100 TraI36 or R-R193Q/Q201R despite affinities 200- and 600-fold lower than R-R193Q/Q201R (data not shown). R-R193Q and R-Q201R have not fully switched specificities as, unlike wild-type F TraI36, they still cleave the R100 oriT oligonucleotide well.
The results of the oligonucleotide cleavage assays suggest that the differences between the binding and cleavage specificities of F and R100 TraI36 reflect differences in both binding and catalysis. The ability of a TraI36 protein to cleave F and R100 oriT oligonucleotides seems to correlate better with the amino acids at 193 and 201 than the relative affinity of the protein for the oligonucleotides. For example, variant F-Q193R/R201Q has 7-fold higher affinity for the R100 oriT site than does F TraI36, and only 4-fold higher affinity than the R-R193Q/Q201R variant, yet cleaves the R100 oligonucleotide almost to completion at 100 nM, whereas the other two proteins generate little product at this concentration. Substitutions that increase the dissociation rate constant of the 5′ cleaved product can make the variant appear more catalytically active because more cleavage product is accumulated. This is due to dissociation becoming more favored relative to the reverse (ligation) reaction (24). Although an enhanced dissociation rate constant could explain the observed differences in the apparent cleavage activities for the F and R100 variants, it is also possible that oligonucleotides bound to a TraI36 protein bearing the R193/Q201 combination are more readily cleaved. This could be a direct influence of these amino acids on the catalytic mechanism, or could be indirect, resulting from their effects on the orientation of other amino acids or bound DNA.
Implications. Our results indicate that the specificity of F and F-like relaxases, highly sequence-specific ssDNA-binding proteins, can be altered with as few as two amino acid substitutions. Similar conversions between naturally occurring binding specificities on substitution of one or a few amino acids have also been reported for several sequence-specific double-stranded (ds) DNA-binding proteins (34–39). Despite this apparent similarity, our relaxase results are distinct from those of dsDNA binding proteins for a number of reasons. First, the stoichiometry of TraI36 binding is 1:1. In contrast, many of the dsDNA-binding proteins studied are oligomeric or are monomers that bind cooperatively to two or more distinct sites. For these proteins, the use of multiple binding units amplifies the effect of any single amino acid substitution. Second, the level of specificity exhibited by F TraI36 suggests that the binding site has a high degree of surface complementarity to the DNA. This complementarity is likely to be highly refined and potentially difficult to alter successfully, especially with few amino acid changes. Third, although relaxase activity is essential to conjugative plasmid transfer, it is not essential for plasmid maintenance. It therefore seems possible that a plasmid could endure a multistep process that alters relaxase recognition and the oriT sequence while remaining viable, even if the intermediate mutational steps left the plasmid unable to transfer. In contrast, it seems less likely that such a process could be tolerated for a DNA-binding protein that regulates an essential cellular process that when disrupted could result in cell death. Fourth, TraI has both ssDNA-binding and -cleavage activities. The particular geometry required by the enzymatic reaction and the constraints that it would put on the architecture of the binding and active site might limit the range of specificities and the manner in which specificity is determined.
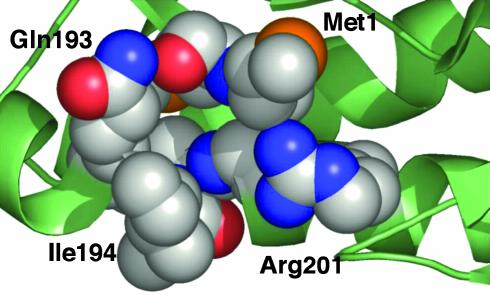
The crystal structure of F TraI36 (40) provides some insight into ssDNA recognition by TraI36. The binding surface of F TraI36 includes a number of pockets or pits ≥4 Å deep and 7–10 Å in diameter, which modeling experiments suggest are capable of accommodating DNA bases (40). Gln-193 and Arg-201 are located on opposite sides of one of these pockets (Fig. 7). Arg-201 also forms part of a second pocket. DNA recognition by TraI36 probably occurs in part through a knob-into-hole fashion, consistent with good surface complementarity and the observed high sequence specificity. This simple model of interaction, however, may be insufficient to account for all of the binding data. We continue attempts at cocrystallization of TraI36 with ssDNA in an effort to gain further insight into the structural basis of their interaction.
Fig. 7.
Amino acids Gln-193 and Arg-201 line pockets within the TraI36-binding surface. The two amino acids are located on opposite sites of a pocket within the DNA-binding surface, as shown by the x-ray crystal structure of F TraI36 (ref. 40; PDB code 1P4D). Shown is a portion of the structure with the main chain shown as a ribbon diagram. The constituent atoms of the pocket are shown as spheres, with carbon in gray, nitrogen in blue, oxygen in red, and sulfur in orange. Side chains are labeled. Also forming the pocket are Gly-197 and the main chain atoms of Met-2, Ala-195, and Phe-196.
We show here that relaxase specificity can be enormously influenced by the amino acids at certain key positions, and substitutions at these positions can alter specificities dramatically. Similar observations have been made for a number of dsDNA-binding proteins. Relaxases differ remarkably from dsDNA-binding proteins, however, in that the key specificity-determining residues of relaxases may be frequently located at pockets within their DNA-binding surface.
Acknowledgments
We thank Dmitri Toptygin for helpful discussions of fluorescence; Dmitri Toptygin, Daniel Isom, and Christopher Larkin for guidance in data collection; and Ludwig Brand, Robert Schleif, and Douglas Fam-brough for use of equipment. This article is based on work funded by National Science Foundation Grant MCB-9733655 and National Institutes of Health Grant GM61017.
Abbreviations: TraI36, 36-kDa relaxase domain of TraI; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; oriT, plasmid origin of transfer; TAMRA, carboxytetramethylrhodamine.
References
- 1.Firth, N., Ippen-Ihler, K. & Skurray, R. A. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol. Press, Washington, DC), 2nd Ed., pp. 2377–2401.
- 2.Frost, L. S., Ippen-Ihler, K. & Skurray, R. A. (1994) Microbiol. Rev. 58, 162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zechner, E. L., de la Cruz, F., Eisenbrandt, R., Grahn, A. M., Koraimann, G., Lanka, E., Muth, G., Pansegrau, W., Thomas, C. M., Wilkins, B. M. & Zatyka, M. (2000) in The Horizontal Gene Pool, ed. Thomas, C. M. (Harwood Academic, Amsterdam), pp. 87–174.
- 4.Carter, J. R. & Porter, R. D. (1991) J. Bacteriol. 173, 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llosa, M., Bolland, S., Grandoso, G. & de la Cruz, F. (1994) J. Bacteriol. 176, 3210–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pansegrau, W., Balzer, D., Kruft, V., Lurz, R. & Lanka, E. (1990) Proc. Natl. Acad. Sci. USA 87, 6555–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furste, J. P., Pansegrau, W., Ziegelin, G., Kroger, M. & Lanka, E. (1989) Proc. Natl. Acad. Sci. USA 86, 1771–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard, M. T., Nelson, W. C. & Matson, S. W. (1995) J. Biol. Chem. 270, 28381–28386. [PubMed] [Google Scholar]
- 9.Pansegrau, W., Schroder, W. & Lanka, E. (1993) Proc. Natl. Acad. Sci. USA 90, 2925–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pansegrau, W., Ziegelin, G. & Lanka, E. (1990) J. Biol. Chem. 265, 10637–10644. [PubMed] [Google Scholar]
- 11.Reygers, U., Wessel, R., Muller, H. & Hoffmann-Berling, H. (1991) EMBO J. 10, 2689–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matson, S. W. & Morton, B. S. (1991) J. Biol. Chem. 266, 16232–16237. [PubMed] [Google Scholar]
- 13.Matson, S. W., Nelson, W. C. & Morton, B. S. (1993) J. Bacteriol. 175, 2599–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llosa, M., Grandoso, G. & de la Cruz, F. (1995) J. Mol. Biol. 246, 54–62. [DOI] [PubMed] [Google Scholar]
- 15.Inamoto, S., Yoshioka, Y. & Ohtsubo, E. (1991) J. Biol. Chem. 266, 10086–92. [PubMed] [Google Scholar]
- 16.Grandoso, G., Avila, P., Cayon, A., Hernando, M. A., Llosa, M. & de la Cruz, F. (2000) J. Mol. Biol. 295, 1163–1172. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda, H. & Ohtsubo, E. (1997) Genes Cells 2, 735–751. [DOI] [PubMed] [Google Scholar]
- 18.Sherman, J. A. & Matson, S. W. (1994) J. Biol. Chem. 269, 26220–26226. [PubMed] [Google Scholar]
- 19.Llosa, M., Grandoso, G., Hernando, M. A. & de la Cruz, F. (1996) J. Mol. Biol. 264, 56–67. [DOI] [PubMed] [Google Scholar]
- 20.Willetts, N. & Maule, J. (1986) Genet. Res. 47, 1–11. [DOI] [PubMed] [Google Scholar]
- 21.Everett, R. & Willetts, N. (1982) EMBO J. 1, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson, R., Taylor, L., Kelly, K., Everett, R. & Willetts, N. (1984) EMBO J. 3, 1175–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inamoto, S., Fukuda, H., Abo, T. & Ohtsubo, E. (1994) J. Biochem. 116, 838–844. [DOI] [PubMed] [Google Scholar]
- 24.Stern, J. C. & Schildbach, J. F. (2001) Biochemistry 40, 11586–11595. [DOI] [PubMed] [Google Scholar]
- 25.Waters, V. L., Hirata, K. H., Pansegrau, W., Lanka, E. & Guiney, D. G. (1991) Proc. Natl. Acad. Sci. USA 88, 1456–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willetts, N. & Maule, J. (1979) Mol. Gen. Genet. 169, 325–336. [DOI] [PubMed] [Google Scholar]
- 27.Street, L. M., Harley, M. J., Stern, J. C., Larkin, C., Williams, S. L., Miller, D. L., Dohm, J. A., Rodgers, M. E. & Schildbach, J. F. (2003) Biochim. Biophys. Acta 1646, 86–99. [DOI] [PubMed] [Google Scholar]
- 28.Fekete, R. A. & Frost, L. S. (2000) J. Bacteriol. 182, 4022–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., Fritsch, E. F. & Maniatis, F. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 30.Harley, M. J., Toptygin, D., Troxler, T. & Schildbach, J. F. (2002) Biochemistry 41, 6460–6468. [DOI] [PubMed] [Google Scholar]
- 31.Toptygin, D. & Brand, L. (1995) Anal. Biochem. 224, 330–338. [DOI] [PubMed] [Google Scholar]
- 32.Toptygin, D. & Brand, L. (1995) Spectrabind User's Guide (The Johns Hopkins Univ., Baltimore, MD).
- 33.Lustig, B. & Jernigan, R. L. (1995) Nucleic Acids Res. 23, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alroy, I. & Freedman, L. P. (1992) Nucleic Acids Res. 20, 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschmied, L., Baumeister, R., Pfleiderer, K. & Hillen, W. (1988) EMBO J. 7, 4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czerny, T. & Busslinger, M. (1995) Mol. Cell. Biol. 15, 2858–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanes, S. D. & Brent, R. (1989) Cell 57, 1275–1283. [DOI] [PubMed] [Google Scholar]
- 38.Raumann, B. E., Knight, K. L. & Sauer, R. T. (1995) Nat. Struct. Biol 2, 1115–1122. [DOI] [PubMed] [Google Scholar]
- 39.Solano, R., Fuertes, A., Sanchez-Pulido, L., Valencia, A. & Paz-Ares, J. (1997) J. Biol. Chem. 272, 2889–2895. [DOI] [PubMed] [Google Scholar]
- 40.Datta, S., Larkin, C. & Schildbach, J. F. (2003) Structure (London), in press. [DOI] [PubMed]