Abstract
Inorganic polyphosphate (poly P), chains of hundreds of phosphate residues linked by “high-energy” bonds as in ATP, has been conserved from prebiotic times in all cells. Poly P is essential for a wide variety of functions in bacteria, including virulence in pathogens. In this study, we observe the unique and many-fold stimulation by poly P in vitro of the protein kinase mTOR (mammalian target of rapamycin). To explore the role of poly P in mammalian cells, a yeast polyphosphatase, PPX1, was inserted into the chromosomes of MCF-7 mammary cancer cells. The transfected cells are markedly deficient in their response to mitogens, such as insulin and amino acids, as seen in their failure to activate mTOR to phosphorylate one of its substrates, PHAS-I (the initiation factor 4E-binding protein). In addition, the transfected cells are severely reduced in their growth in a serum-free medium. On the basis of these findings, we suggest that poly P (and/or PPX1) serves as a regulatory factor in the activation of mTOR in the proliferative signaling pathways of animal cells.
Inorganic polyphosphate (poly P), linear polymers of many hundreds of phosphate (Pi) residues linked by the “high-energy” phosphoanhydride bonds found in ATP, is present in all bacterial, fungal, plant, and animal cells (1). Escherichia coli poly P levels increase 100- to 1,000-fold in response to some stresses, such as the “starvation” in a nutrient downshift from a rich to a minimal medium. The increase in poly P helps the bacteria respond to the lack of amino acids by activating Lon protease to provide them (2). Although poly P is ubiquitous in mammalian cells and tissues (3), its biological role(s) in these and other eukaryotic organisms has not been defined.
In mammalian cells, the protein kinase mTOR (mammalian target of rapamycin) integrates information from mitogenic signals, energy levels, and nutrient sufficiency (e.g., amino acids) to control cellular growth and proliferation (Fig. 1 Left) (4, 5). This enzyme (also called FRAP or RAFT) belongs to the family of phosphatidylinositol kinase-related kinases (PIKK) and regulates cell growth in large part by its ability to phosphorylate two substrates: ribosomal S6 kinase and PHAS-I, the eukaryotic translation initiation factor 4E-binding protein (4–6). Phosphorylation of PHAS-I by mTOR releases the mRNA cap-binding protein, eIF4E, which is involved in initiation of translation of a subset of mRNAs important for cell growth and proliferation (7). Phosphorylation of the S6 kinase and its substrate, ribosomal protein S6, has also been proposed to stimulate translation of a subset of mRNAs containing a 5′-terminal oligopolypyrimidine tract (8). Thus, activation of mTOR stimulates protein synthesis in mammalian cells, whereas in nutrient-deprived conditions (e.g., insufficient amino acids, mitogens) mTOR activity is decreased, thereby limiting protein synthesis and cellular growth.
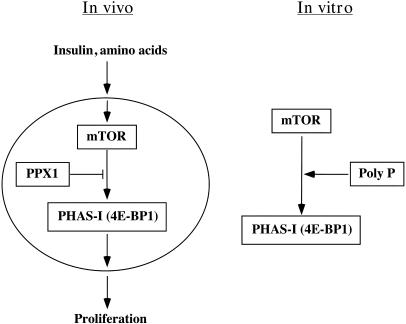
Fig. 1.
Scheme for poly P role in the mTOR signaling pathway. Only the portions of the known pathways relevant to the results presented in this work are shown. Poly P is proposed to act as a cofactor in the insulin and amino acid activation of mTOR function. Other aspects are discussed in the text. See ref. 4 for a recent review on TOR.
We now find that poly P is likely a regulator of mTOR activity both in vitro and in vivo. Poly P chains of lengths 15–750, but not as short as 5, Pi residues are potent stimulators of isolated mTOR kinase activity. Moreover, expression of the yeast exopolyphosphatase PPX1 gene (9) in the human breast carcinoma cell line MCF-7 greatly inhibits the ability of a mitogen, insulin, or amino acids to stimulate the phosphorylation of the endogenous mTOR substrate, PHAS-I. Further, expression of PPX1 inhibited the proliferation of these cells in a serum-free medium. Thus, we suggest a role for poly P in the regulation of mTOR, and demonstrate that a regulatory role for poly P in responding to a stress, such as nutritional deficiency, may be conserved from prokaryotes to mammalian cells.
Materials and Methods
Materials. [γ-32P]ATP (3,000 Ci/mmol; 1 Ci = 37 GBq) was purchased from Perkin–Elmer Life Sciences, porcine insulin was from Roche Molecular Biochemicals, the MEM amino acid mixture (50×) was from Cellgro (Herndon, VA), and the bicinchoninic acid protein determination kit was from Pierce. Poly P type 5, type 15, type 35, and type 65, heterogeneous mixtures of chain lengths containing P5, P15, P35, and P65, respectively, were from Sigma. P750 is a relatively monodisperse P700–P800 product synthesized with purified E. coli PPK1, as described (10), except that it was purified by two sequential 2-butanol precipitations instead of cesium chloride cushion centrifugation. Anti-myc 9E-10 antibody was from Calbiochem. Phosphospecific antibodies to the Ser-2448 and -2481 sites in mTOR and Ser-65 in PHAS-I were from Cell Signaling Technology (Beverly, MA). An anti-mTOR antiserum was a kind gift of Jie Chen (University of Illinois at Urbana–Champaign, Urbana), and the anti-PHAS-I antiserum was a gift of John Lawrence (University of Virginia, Charlottesville). The anti-Flag M2 affinity gel and all other chemicals were also from Sigma or as indicated.
Construction of an Expression Plasmid for PPX1. The Saccharomyces cerevisiae gene PPX1, which encodes a major exopolyphosphatase (9), was cloned into the mammalian expression vector pcDNA3.1+ (Invitrogen) previously modified by the addition of a myc tag sequence and altered restriction endonuclease sites near the 5′ end of the multiple cloning site downstream of the cytomegalovirus promoter (11). PPX1 was subcloned from pTrcPPX1 (9) by standard PCR using Taq Core Polymerase (Qiagen, Valencia, CA) and the following primers: forward 5′TCGCCTTTGAGAAAGACGGTT3′ and reverse 5′CTCCCTTCACTCTTCCAGGTT3′, with appropriate restriction sites on the 5′ ends, followed by standard ligation reactions with T4 DNA Ligase (New England Biolabs). The construction was verified by restriction endonuclease analysis.
Cell Culture and Cell Lines. CHO-T cells (12) and MCF-7 cells were cultured in Ham's F-12 medium (Cellgro) and DMEM/F-12 (GIBCO/Invitrogen), respectively, supplemented with 10% FCS (Gemini Biological Products, Calabasas, CA), streptomycin, and penicillin. To generate the transfected cell lines used in the present study, the lipid method was employed, using FuGENE 6 Transfection Reagent (Roche Molecular Biochemicals) with either the control pcDNA3.1 plasmids or the plasmid encoding the myc-tagged PPX1 described above. Transfections were similarly performed with FLAG-tagged wild-type mTOR or FLAG-tagged kinase-dead mTOR (with mutation D2357E; ref. 13) (gifts of J. Chen and S. Schreiber, Harvard University, Cambridge, MA). The cells were selected with 1 mg/ml G418 and cloned, and the isolated clones were screened for expression of the plasmids.
Immunoprecipitation of mTOR and in Vitro Kinase Assays. CHO-T cells stably expressing the indicated mTOR were washed and lysed in ice-cold Nonidet P-40 lysis buffer (1% Nonidet P-40/150 mM NaCl/20 mM Tris, pH 8.0/1mMDTT/10% glycerol/1mM phenylmethanesulfonyl fluoride/0.5 mM Na3VO4/100 nM okadaic acid/10 μg/ml aprotinin/10 μg/ml leupeptin). The cleared supernatants were incubated with anti-Flag agarose for 4–6 h at 4°C. The beads and their bound proteins were then washed once with lysis buffer containing 0.5 M NaCl, twice with buffer containing 50 mM Tris (pH 7.4) and 150 mM NaCl, and once with 1× kinase buffer (10 mM Hepes, pH 7.4/50 mM β-glycerophosphate/10 MnCl2/50 mM NaCl). The inclusion of detergent and high salt in the washes decreases most mTOR-associated proteins, including raptor (14). The mTOR kinase assays were performed in a total reaction volume of 30 μl containing 300 μM ATP, 1 μg of PHAS-I (Calbiochem), and 10 μCi of [γ-32P]ATP, with or without poly P, for 30 min at 30°C. In some experiments, the poly P was first incubated with purified yeast PPX1 (9). Reactions were terminated by the addition of 10 μl of 4× sample buffer and boiling. Samples were analyzed by 3–10% SDS/PAGE. Phosphorylated proteins were visualized by autoradiography and quantified with a PhosphorImager and imagequant software (Amersham Biosciences) in arbitrary units.
PPX1 Activity Assays. MCF-7 cells transfected with either the control pcDNA3.1 vector or the plasmid expressing PPX1 were grown to confluency, washed three times with PBS, and lysed in 500 μl of 20 mM Tris·HCl (pH 7.4)/10% glycerol/1% Nonidet P-40/1 mM phenylmethanesulfonyl fluoride. The lysates were sonicated four times for 20–30 sec at 100% amplitude in a Sonics Vibra Cell cup horn sonicator with 30- to 60-sec cooling intervals on ice. Each lysate (10 μl) was incubated in a 15-μl reaction mixture with 1× ScPPX1 buffer (15) and 15 nmol of [32P] P750 (Pi residues) at 37°C. After 1, 5, and 15 min, a sample (3 μl) of the reaction was stopped by addition of EDTA to a final concentration of 50 mM, spotted onto a polyethyleneimine-cellulose F TLC plate (EM Science), and run in a 0.4 M lithium chloride/1.0 M formic acid solvent system. The TLC plate was then exposed to a PhosphorImager plate that was scanned in an Amersham Biosciences Typhoon 8600; the image was analyzed by using imagequant volume analysis software. One unit corresponds to the release of 1 pmol of Pi per minute at 37°C (9).
In Vivo Assays of mTOR Activity. MCF-7 cells transfected with either control pcDNA3.1 or its PPX1-expressing derivative were first grown to 50% confluency in 10-cm dishes. The medium was then removed and replaced with Hanks' balanced salt solution (HBSS) (Invitrogen/GIBCO). After 44 h, the cells were either left as is or stimulated with a mixture of amino acids or insulin, as indicated. Thirty minutes later the cells were lysed as described above and the lysates were analyzed by SDS/PAGE and immunoblotting. Blots were developed with horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences) and enhanced chemiluminescence substrate (Pierce). They were then scanned on a Kodak Image Station 440CF and quantified in arbitrary units.
Measurement of Cell Proliferation. MCF-7 cells were cultured in six-well plates in DMEM/F-12 with or without 10% FCS. On the indicated days, cells were rinsed, trypsinized, pelleted by centrifugation, stained with trypan-blue, and counted by light microscopy.
Results
Poly P Stimulates mTOR Kinase Activity in Vitro. CHO-T cells stably expressing Flag-tagged mTOR were either untreated or maximally stimulated with insulin plus amino acids. The mTOR was immunoprecipitated from lysates and assayed for kinase activity on its substrate, PHAS-I (16). Although these conditions stimulated an in vivo increase in endogenous PHAS-I phosphorylation (data not shown), no significant in vitro increase was observed with mTOR kinase obtained from insulin/amino acid-treated cells over that observed with nontreated cells (Fig. 2). Thus, the stimulation of mTOR kinase activity in intact cells is apparently not due to a stable modification (i.e., phosphorylation) of the enzyme but to factors responsive to insulin and/or amino acid signals.
Fig. 2.
Stimulation of mTOR kinase by in vitro versus in vivo treatments. CHO-T cells stably expressing a Flag-tagged mTOR were starved for serum and amino acids for 16 h and then either left untreated or stimulated with a mixture of amino acids (at the concentration in MEM medium) plus 100 nM insulin for 30 min. The mTOR was immunoprecipitated from the lysates of 3 × 106 cells and assayed for kinase activity by using isolated PHAS-I as substrate. Duplicate samples were assayed in the presence of 0.5 μM poly P65. (Upper) Autoradiogram showing the phosphorylation of the PHAS-I substrate. Results were quantitated and expressed as the fold-increase over the control, nontreated cells.
Poly P serves as a cofactor to activate the Lon protease in E. coli under nutrient-deprived conditions (2). Similarly, poly P might serve as a cofactor for mTOR, a mammalian sensor of the nutritional environment of the cell. Addition of poly P65 (0.5 μM) stimulated the phosphorylation of PHAS-I by mTOR from either starved or insulin/amino acid-treated cells 3.5-fold (Fig. 2).
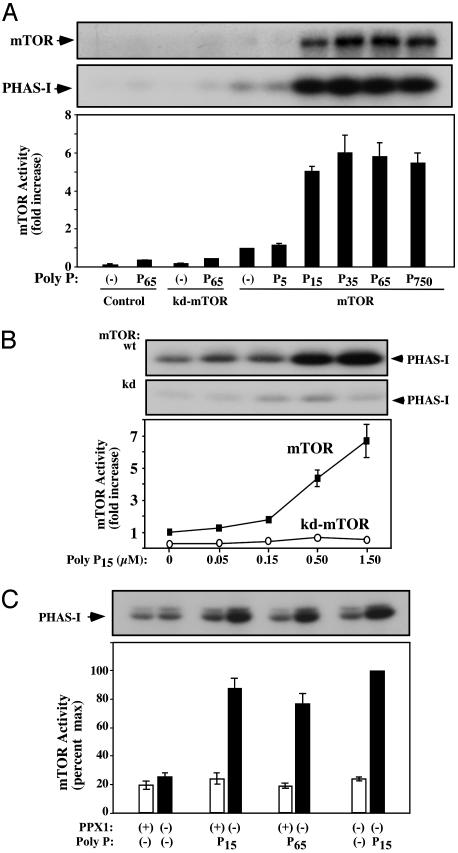
Poly P over a range of long chain lengths (P15, P35, P65, and P750) at 0.5 μM stimulated the mTOR phosphorylation of PHAS-I, but shorter chains (e.g., P5) did not (Fig. 3A). Concomitant with the increase in PHAS-I phosphorylation, poly P also stimulated the autophosphorylation of mTOR (Fig. 3A Top). Western blots of these samples with antibodies that bind mTOR at a specific phosphorylated residue (Ser-2481) confirmed that the poly P-stimulated increase in mTOR phosphorylation occurred on Ser-2481, a known mTOR autophosphorylation site (data not shown) (17). However, no increase in phosphorylation was observed at Ser-2448 (data not shown), a site normally phosphorylated by an exogenous kinase (18). Thus, under the conditions tested, poly P appears to stimulate the intrinsic activity of mTOR and not that of an associated kinase. The specificity of the poly P stimulation of mTOR is also supported by the finding that DNA-dependent protein kinase, a homologue in the PIKK family (19), is not activated by poly P in vitro (L. G. DeFazio and G. Chu, personal communication).
Fig. 3.
Poly P stimulation of mTOR kinase activity. (A) Effect of poly P chain length. Either wild-type mTOR (mTOR) or kinase-dead mTOR (kd) were immunoprecipitated and assayed for kinase activity in the presence or absence of 0.5 μM of the indicated poly P. Results of three experiments were averaged and expressed as the fold-increase ± SE over the nontreated wild-type mTOR control; Top and Middle show autoradiograms of representative experiments with an arrow indicating the position of the mTOR and PHAS-I proteins. (B) Concentration dependence of the poly P stimulation. Wild-type mTOR (mTOR) or kinase-dead (kd)-mTOR were immunoprecipitated and assayed for kinase activity in the presence of the indicated concentrations of poly P15.(C) Hydrolysis of poly P eliminates mTOR activation. Assays were performed as above, except that the poly P was first incubated as indicated with a purified preparation of the yeast exopolyphosphatase, PPX1.
To further test whether these stimulating effects of poly P could be due to another tightly associated kinase, a comparable amount of a kd-mTOR was immunoprecipitated (17). This mutant enzyme exhibited almost no exogenous substrate or autophosphorylation kinase activity, even in the presence of activating levels of poly P65 (Fig. 3A). Clearly it is the intrinsic mTOR activity that is stimulated by poly P. To confirm these poly P effects, the wild-type mTOR and the kd-mTOR were immunoprecipitated and tested for kinase activity at different P15 concentrations. Wild type, but not kd-mTOR, was stimulated in a dose-dependent manner with an almost 7-fold stimulation at 1.5 μM P15 (Fig. 3B).
To verify that the increase in enzymatic activity observed with the different poly P preparations was not due to a contaminant in these preparations, the poly P was first treated with a purified yeast exopolyphosphatase PPX1 sufficient to digest all of the poly P in the reaction (9, 15). The stimulations observed with either P15 or P65 were completely eliminated by such prior PPX1 digestions (Fig. 3C).
Autophosphorylation of mTOR Minimally Stimulates Its PHAS-I Kinase Activity. To determine whether poly P autophosphorylation of mTOR might be responsible for its stimulation of PHAS-I phosphorylation, immunoprecipitated mTOR was autophosphorylated in either the presence or absence of poly P and ATP, and then washed and tested for its kinase activity on PHAS-I. Whereas prior incubation with either ATP or poly P alone caused no significant increase in mTOR activity (Fig. 4), a 50% increase in activity was consistently observed with mTOR first incubated with ATP plus poly P (Fig. 4), conditions which allow maximal autophosphorylation (Fig. 3A). However, this activation of mTOR kinase activity was far lower than the 5-fold increase in activity seen when poly P was included in the reaction with PHAS-I (Fig. 4). Thus, the bulk of the poly P-stimulated kinase activity was not due to autophosphorylation and prior activation of mTOR. These results, together with the finding that ATP does not compete with poly P in its stimulatory activity (data not shown), indicate that poly P regulates mTOR in an allosteric manner.
Fig. 4.
Autophosphorylation of mTOR minimally stimulates its PHAS-I kinase activity. Flag-tagged wild-type mTOR (wt) and kinase-dead mTOR (kd) were immunoprecipitated and allowed to autophosphorylate in the presence or absence of ATP and poly P, as indicated. The mTOR (still bound to the beads) was washed twice with ice-cold PBS and then assayed for kinase activity. Radioactivity incorporated into the PHAS-I band in three experiments was quantitated, and the averaged mTOR activity was expressed as the fold-increase ± SE over the control wild-type mTOR. (Upper) Autoradiogram showing the phosphorylation of the PHAS-I substrate.
Polyphosphatase PPX1 Inhibits mTOR Activation in Human Breast Cancer Cells. MCF-7 cells, a breast carcinoma cell line, were stably transfected with pcDN3.1 encoding myc-tagged yeast exopolyphosphatase PPX1 (9). This enzyme hydrolyzes poly P of varying chain lengths but does not act on ATP or inorganic pyrophosphate (15). Two independent cell clones were derived that expressed the myc-tagged PPX1, as demonstrated by the presence of an appropriately sized 45-kDa band detected with anti-myc antibodies (data not shown). These cells expressed ≈1,500 PPX1 units/mg of total cell protein, whereas control cells transfected with the vector plasmid, pcDNA3.1, showed no comparable myc-tagged band and had no detectable PPX1 activity (<0.1%).
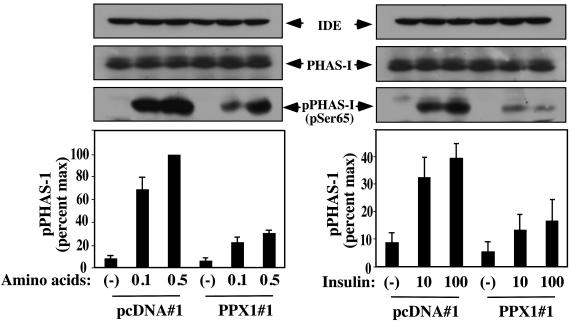
These stably transfected cells were used to determine whether PPX1 expression would affect the in vivo mTOR enzymatic activity. Cells in nutrient-deprived medium (HBSS) were treated with either amino acids or insulin and lysed; lysates were tested for the presence of PHAS-I phosphorylated on a specific residue (Ser-65) as determined by immunoblotting. The control cells (pcDNA#1) exhibited the anticipated amino acid- and insulin-stimulated increases in PHAS-I phosphorylation (Fig. 5), but the phosphorylation was significantly reduced in the PPX1-expressing cells (PPX1#1) (Fig. 5). Immunoblot controls for total PHAS-I as well as an extraneous protein (IDE) verified that these cells contained comparable amounts of both proteins (Fig. 5), indicating that PPX1 expression did not change their levels. In addition, PPX1 expression did not effect the insulin stimulation of Akt kinase phosphorylation (data not shown), demonstrating that the effect of PPX1 on mTOR activity was kinase-specific.
Fig. 5.
Polyphosphatase PPX1 reduces insulin and amino acid activation of mTOR activity in vivo. Serum- and amino acid-starved MCF-7 cells, either the pcDNA3.1 transfected (pcDNA#1) or PPX1-expressing (PPX1#1) cells, were incubated for 30 min with either 0.1 or 0.5 of the normal levels of amino acids in MEM or 10 nM or 100 nM insulin, as indicated. Cell lysates were analyzed by immunoblotting for an extraneous protein (IDE), total PHAS-I levels, or PHAS-I that was phosphorylated on Ser-65. (Upper) Western blots of a representative experiment with arrows indicating the positions of IDE, total PHAS-I protein, and phosphorylated PHAS-I. The amount of pPHAS-I was quantitated, and shown are the averages of three experiments ± SE expressed as a percent of the maximally stimulated cells.
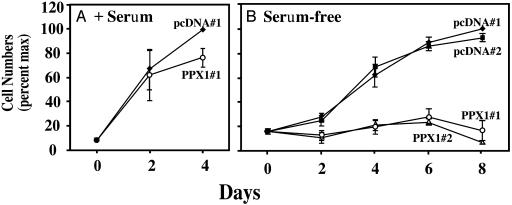
In as much as mTOR regulates cellular proliferation (4, 5), the growth rates of the control and PPX1-expressing cells were examined. Whereas a small difference in the proliferation of these cells in complete, serum-containing medium was observed (Fig. 6A), the growth of the PPX1-expressing cells in serum-free medium was dramatically decreased (Fig. 6B); this effect was observed in both of the independently derived clones of PPX1-expressing cells (PPX1#1 and PPX1#2; Fig. 6B).
Fig. 6.
Polyphosphatase PPX1 inhibits the proliferative pathway of MCF-7 cells in serum-free medium. MCF-7 cells transfected with either vector plasmid (clones pcDNA#1 and pcDNA#2) or PPX1 (clones PPX1#1 and PPX1#2) were grown in DMEM/F-12 medium with (A) or without (B) 10% FCS. The numbers of viable cells of each type in each of three experiments were averaged, and the numbers are expressed as percent of the maximum ± SE. In A, the cells reached confluence after 4 days.
Discussion
TOR is a protein phosphotransferase whose role in cell growth and proliferation has been conserved from yeast to mammals (4, 5). TOR activity responds to levels of nutrients and growth factors in the environment and regulates protein synthesis and cellular proliferation. An inhibitor of TOR function, rapamycin, is a candidate antitumor drug that counteracts the increased levels of TOR activity in many tumors and inhibits neoplastic proliferation (20–22).
Extensive studies in a variety of organisms have documented two key downstream targets of TOR (4, 5). PHAS-I (also called eIF4E-binding protein) in its unphosphorylated state sequesters eIF4E, an mRNA cap-binding protein in the eIF4G elongation complex. PHAS-I phosphorylation by mTOR releases eIF4E, stimulating both global translation as well as that of a particular set of proteins involved in cell growth and proliferation. This appears to be a crucial regulatory mechanism, in as much as overexpression of eIF4E alone can stimulate the transformation of rodent fibroblasts (23). The other major mTOR substrate is the kinase for the 40S ribosomal S6 protein (p70S6 kinase), phosphorylation of which activates its enzymatic activity. Phosphorylation of S6 reportedly enhances the translation of mRNAs containing a 5′-terminal oligopolypyrimidine tract, although this has recently been questioned (24).
It is therefore of key interest to determine how various growth factors and nutrients regulate mTOR enzymatic activity in vivo. One signaling cascade implicated in this process is the phosphatidylinositol triphosphate/Akt pathway (5, 25). Mammalian TOR was observed to be phosphorylated by Akt on Ser-2448 (18); however, a mutation of this site had little effect on the in vivo regulation of mTOR (26). In addition, the role of phosphorylation of mTOR, either autocatalyzed or as a kinase substrate, in its stimulation is not clear because the enzyme isolated from cells treated to activate this pathway shows at most a 2-fold activation of its substrate kinase activity (27). Efforts to explain mTOR regulation in vivo by growth factors and nutrients have suggested several potential regulatory cellular components, such as phosphatidic acid (28) and ATP (29). Recently, regulation by mTOR binding to a scaffold protein termed Raptor (14, 30) and/or binding to a complex of two proteins designated TSC1 and TSC2 (31) have also been shown. It has also been proposed that the primary in vivo regulation of mTOR is not through a change in its intrinsic enzymatic activity but through its ability to regulate an associated phosphatase activity (32).
Here we demonstrate that inorganic polyphosphate (poly P) regulates the activity and function of mTOR, a eukaryotic kinase involved in cellular proliferation. Poly P is ubiquitous in nature, being found in all organisms from bacteria to animals (1). In E. coli, levels of poly P respond to stresses such as nutritional downshifts (1). After a shift to an amino acid-deprived environment, poly P stimulates ribosomal protein degradation by binding to and activating the Lon protease, thereby supplying amino acids needed to respond to starvation (2). The present studies show that poly P also stimulates the in vitro activity of mTOR to both autophosphorylate as well as to phosphorylate its in vivo substrate, PHAS-I, from 3.5- to 7-fold (Fig. 1). This activation occurred at concentrations of poly P (0.15–1.5 μM) found in mammalian cells§ (3) and with poly P chain lengths of 15–750 phosphate residues. mTOR autophosphorylation in vitro in the presence of poly P and ATP induced a 50% increase in enzyme activity, similar to those previously reported after cells were treated with growth factors and nutrients (27).
The ability of poly P to activate mTOR in vitro was corroborated in vivo with a human carcinoma cell line (MCF-7) genetically manipulated to express the yeast exopolyphosphatase PPX1 gene (9). This enzyme degrades poly P of varying lengths to Pi but does not act on ATP or pyrophosphate (9). In contrast to control cells transfected with empty plasmid, these cells exhibited a greatly reduced response to either amino acid- or insulin-stimulated phosphorylation of PHAS-I. Moreover, PPX1-expressing cells failed to grow in a serum-free medium, although their growth in serum-containing medium was not dramatically affected. This growth defect was not due to changes in ATP levels (data not shown), nor was the ability of insulin to stimulate Akt phosphorylation compromised in the PPX1-expressing cells (data not shown). However, consistent with a role for mTOR in determining cell size (8), the PPX1-expressing cells were ≈25% smaller than the control cells in serum-free media (data not shown). Thus, under the conditions examined, the exopolyphosphatase effect appears to be pathway-specific.
In ongoing studies, no gross changes in total poly P were observed in crude lysates of PPX1-expressing cells, but further examinations need to be made of the possible influence of PPX1 on the distribution of poly P chain lengths and their localization in subcellular compartments. It is also possible that the expressed PPX1 interfered with mTOR signaling by a mechanism other than by its action as a polyphosphatase. PPX1 is a member of the DHH family of nucleases and phosphatases and, as such, it is highly homologous to the Prune protein (33). Prune has been implicated in the regulation of tumor metastasis in humans; increased levels of Prune have been reported in human sarcomas and breast cancers (34). In contrast to the PPX1 growth defect described here, a previous study showed that Prune expression resulted in cellular proliferation (34), suggesting different functions for the two proteins. However, it is not known whether Prune has polyphosphatase activity. In addition, transfection of MCF-7 cells with a site-directed polyphosphatase activity mutant of PPX1 will help determine whether this activity is responsible for the in vivo observed effects of PPX1.
In brief, our studies demonstrate that poly P stimulates mTOR function in vitro and may activate it in vivo (Fig. 1). Although poly P had previously been identified in a variety of mammalian cells and tissues (3), its role has yet to be defined. Based on the profound effects of poly P in regulating signaling in bacteria, it seems plausible that it may be used in responses to environmental factors in animal cells as well. Not only has poly P itself been conserved in biotic evolution, but at least one of its functions, as a regulator of basic cellular processes in response to growth conditions or stresses, has been conserved as well.
Acknowledgments
We thank Drs. J. Chen and S. Schreiber for the gifts of the mTOR plasmids, Dr. Jie Chen for the antiserum to mTOR, and Dr. John Lawrence for the antiserum to PHAS-I. This work was supported in part by National Institutes of Health Grants DK34926 (to R.A.R.) and GM007581 (to A.K.) and National Cancer Institute Training Grant PHS CA09302 (to J.F.).
Abbreviations: mTOR, mammalian TOR; kd, kinase dead; Pi, phosphate; poly Pn, polyphosphate where n is the chain length of Pi residues.
Footnotes
In this article, poly P concentrations are all expressed in terms of molecules of intact poly P rather than phosphate or Pi content.
References
- 1.Kornberg, A., Rao, N. N. & Ault-Riché, D. (1999) Annu. Rev. Biochem. 68, 89–125. [DOI] [PubMed] [Google Scholar]
- 2.Kuroda, A., Nomura, K., Ohtomo, R., Kato, J., Ikeda, T., Takiguchi, N., Ohtake, H. & Kornberg, A. (2001) Science 293, 705–708. [DOI] [PubMed] [Google Scholar]
- 3.Kumble, K. D. & Kornberg, A. (1995) J. Biol. Chem. 270, 5818–5822. [DOI] [PubMed] [Google Scholar]
- 4.Jacinto, E. & Hall, M. N. (2003) Nat. Rev. Mol. Cell Biol. 4, 117–126. [DOI] [PubMed] [Google Scholar]
- 5.Martin, K. A. & Blenis, J. (2002) Adv. Cancer Res. 86, 1–39. [DOI] [PubMed] [Google Scholar]
- 6.Fingar, D. C., Salama, S., Tsou, C., Harlow, E. & Blenis, J. (2002) Genes Dev. 16, 1472–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miron, M., Verdu, J., Lachance, P. E., Birnbaum, M. J., Lasko, P. F. & Sonenberg, N. (2001) Nat. Cell Biol. 3, 596–601. [DOI] [PubMed] [Google Scholar]
- 8.Kozma, S. C. & Thomas, G. (2002) BioEssays 24, 65–71. [DOI] [PubMed] [Google Scholar]
- 9.Wurst, H., Shiba, T. & Kornberg, A. (1995) J. Bacteriol. 177, 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ault-Riché, D., Fraley, C. D., Tzeng, C. M. & Kornberg, A. (1998) J. Bacteriol. 180, 1841–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovacina, K. S., Park, G. Y., Bae, S. S., Guzzetta, A. W., Schaefer, E., Birnbaum, M. J. & Roth, R. A. (2003) J. Biol. Chem. 278, 10189–10194. [DOI] [PubMed] [Google Scholar]
- 12.Ellis, L., Clauser, E., Morgan, D. O., Edery, M., Roth, R. A. & Rutter, W. J. (1986) Cell 45, 721–732. [DOI] [PubMed] [Google Scholar]
- 13.Brown, E. J., Albers, M. W., Shin, T. B., Ichikawa, K., Keith, C. T., Lane, W. S. & Schreiber, S. L. (1994) Nature 369, 756–758. [DOI] [PubMed] [Google Scholar]
- 14.Hara, K., Maruki, Y., Long, X., Yoshino, K., Oshiro, N., Hidayat, S., Tokunaga, C., Avruch, J. & Yonezawa, K. (2002) Cell 110, 177–189. [DOI] [PubMed] [Google Scholar]
- 15.Wurst, H. & Kornberg, A. (1994) J. Biol. Chem. 269, 10996–11001. [PubMed] [Google Scholar]
- 16.Brunn, G. J., Hudson, C. C., Sekulic, A., Williams, J. M., Hosoi, H., Houghton, P. J., Lawrence, J. C., Jr., & Abraham, R. T. (1997) Science 277, 99–101. [DOI] [PubMed] [Google Scholar]
- 17.Peterson, R. T., Beal, P. A., Comb, M. J. & Schreiber, S. L. (2000) J. Biol. Chem. 275, 7416–7423. [DOI] [PubMed] [Google Scholar]
- 18.Nave, B. T., Ouwens, M., Withers, D. J., Alessi, D. R. & Shepherd, P. R. (1999) Biochem. J. 344, 427–431. [PMC free article] [PubMed] [Google Scholar]
- 19.DeFazio, L. G., Stansel, R. M., Griffith, J. D. & Chu, G. (2002) EMBO J. 21, 3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neshat, M. S., Mellinghoff, I. K., Tran, C., Stiles, B., Thomas, G., Petersen, R., Frost, P., Gibbons, J. J., Wu, H. & Sawyers, C. L. (2001) Proc. Natl. Acad. Sci. USA 98, 10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podsypanina, K., Lee, R. T., Politis, C., Hennessy, I., Crane, A., Puc, J., Neshat, M., Wang, H., Yang, L., Gibbons, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10320–10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hidalgo, M. & Rowinsky, E. K. (2000) Oncogene 19, 6680–6686. [DOI] [PubMed] [Google Scholar]
- 23.Lazaris-Karatzas, A., Montine, K. S. & Sonenberg, N. (1990) Nature 345, 544–547. [DOI] [PubMed] [Google Scholar]
- 24.Tang, H., Hornstein, E., Stolovich, M., Levy, G., Livingstone, M., Templeton, D., Avruch, J. & Meyuhas, O. (2001) Mol. Cell. Biol. 21, 8671–8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohn, A. D., Barthel, A., Kovacina, K. S., Boge, A., Wallach, B., Summers, S. A., Birnbaum, M. J., Scott, P. H., Lawrence, J. C., Jr., & Roth, R. A. (1998) J. Biol. Chem. 273, 11937–11943. [DOI] [PubMed] [Google Scholar]
- 26.Sekulic, A., Hudson, C. C., Homme, J. L., Yin, P., Otterness, D. M., Karnitz, L. M. & Abraham, R. T. (2000) Cancer Res. 60, 3504–3513. [PubMed] [Google Scholar]
- 27.Scott, P. H., Brunn, G. J., Kohn, A. D., Roth, R. A. & Lawrence, J. C., Jr. (1998) Proc. Natl. Acad. Sci. USA 95, 7772–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang, Y., Vilella-Bach, M., Bachmann, R., Flanigan, A. & Chen, J. (2001) Science 294, 1942–1945. [DOI] [PubMed] [Google Scholar]
- 29.Dennis, P. B., Jaeschke, A., Saitoh, M., Fowler, B., Kozma, S. C. & Thomas, G. (2001) Science 294, 1102–1105. [DOI] [PubMed] [Google Scholar]
- 30.Kim, D. H., Sarbassov, D. D., Ali, S. M., King, J. E., Latek, R. R., Erdjument-Bromage, H., Tempst, P. & Sabatini, D. M. (2002) Cell 110, 163–175. [DOI] [PubMed] [Google Scholar]
- 31.Tee, A. R., Fingar, D. C., Manning, B. D., Kwiatkowski, D. J., Cantley, L. C. & Blenis, J. (2002) Proc. Natl. Acad. Sci. USA 99, 13571–13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson, R. T., Desai, B. N., Hardwick, J. S. & Schreiber, S. L. (1999) Proc. Natl. Acad. Sci. USA 96, 4438–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aravind, L. & Koonin, E. V. (1998) Trends Biochem. Sci. 23, 17–19. [DOI] [PubMed] [Google Scholar]
- 34.Forus, A., D'Angelo, A., Henriksen, J., Merla, G., Maelandsmo, G. M., Florenes, V. A., Olivieri, S., Bjerkehagen, B., Meza-Zepeda, L. A., del Vecchio Blanco, F., et al. (2001) Oncogene 20, 6881–6890. [DOI] [PubMed] [Google Scholar]