Abstract
Forkhead-associated (FHA) domains are phosphoprotein-binding modules found in diverse signaling proteins that bind partners phosphorylated on threonine or serine. Kinase-associated protein phosphatase from Arabidopsis employs its FHA domain for negative regulation of receptor-like kinase signaling pathways, which are important in plant development. The solution structure of the free state of kinase-interacting FHA domain (KI-FHA) of kinase-associated protein phosphatase has been determined with high precision and accuracy using residual dipolar couplings. KI-FHA is a sandwich of a five-stranded mixed β-sheet with a six-stranded antiparallel β-sheet. Despite homology only in the recognition loops, this fold is shared with FHA domains from checkpoint proteins from yeast and humans, as well as with nonhomologous MH2 domains of Smad tumor suppressors. A shared pattern of hydrophobicity throughout FHA domains and Smad MH2 domains may stabilize the core of the β-sandwich. Evolutionary trace analysis of FHA domains suggests class-specific residues in the recognition loops that could tune their phosphoprotein-binding specificity. This surface agrees with that of KI-FHA in contact with a phosphothreonine peptide ligand. Evolutionary trace analysis also predicts an unexpected swath of class-specific residues on another face of FHA domains. Protein interactions with these faces may affect assembly of transmembrane signaling complexes in plants, and in other FHA domain-containing assemblies.
Signal transducing Ser/Thr receptor-like protein kinases (RLKs) in plants, numbering 417 in Arabidopsis, were discovered based on their sequence similarity to receptor tyrosine kinases (1). RLKs play crucial roles in self-incompatibility, hormonal signaling, nodulation, abscission, systemic wound responses, and disease resistance in plants (1). The best characterized signaling component downstream of RLKs is kinase-associated protein phosphatase (KAPP). It has an N-terminal type I membrane anchor, a kinase interaction domain, and a protein phosphatase type 2C (PP2C) catalytic domain (2). KAPP interacts with Arabidopsis RLKs, which include HAESA (formerly RLK5) (2), RLK4 (3), TMK1 (3), CLAVATA1 (4), WAK1 (5), FLS2 (6), SERK1 (7), BRI1, and BAK1 (J. Li, J. Wen, and J.C.W., unpublished work). KAPP is thought to bind phosphorylated serine or threonine of the kinase domain of CLAVATA1 to attenuate activation of CLAVATA1 through the PP2C activity of KAPP (2, 4, 8). The kinase interaction domain of KAPP is found on a 239-residue fragment spanning residues 98–336 (2). The minimal kinase interaction domain comprises the 119 amino acids from residue 180 to 298 that contains the FHA motif (9); this domain is designated hereafter as KI-FHA. Conserved residues in the FHA motif, i.e., Gly-211, Ser-226, His-229, and Asn-250 are essential for KI-FHA to bind phosphorylated RLKs (9).
FHA motifs were suggested to be 55–75 residues long, with a glycine, arginine, serine, histidine, and asparagine conserved among three short blocks (10). The FHA motif is found in a number of signaling proteins and transcription factors from a wide range of organisms, including yeast, plants, humans, and a few bacteria (10). Biochemical and structural studies demonstrated that FHA domains are modular phosphoprotein binding domains (9, 11, 12) comprising 120–140 residues (13, 14). Most FHA domains have been localized to the nucleus where they influence transcription, DNA repair, and cell-cycle progression (15). By contrast, Arabidopsis KAPP lacks a nuclear localization signal and is a transmembrane component of the plasma membrane (2). FHA domains serve as modules in multidomain proteins in phosphorylation-dependent localization of signaling molecules through phosphothreonine (pThr)- or phosphoserine (pSer)-mediated protein–protein interactions (15).
Structures of FHA domains from yeast RAD53 (13, 14, 16, 17), its human ortholog, Chk2 (18), and human CHFr (19) were reported. Sequence identity of <20% prevents homology modeling of distantly related KI-FHA from KAPP in plants. An atomic-resolution structure of KI-FHA from KAPP is essential to understand and possibly engineer protein interactions in RLK-dependent signaling in plants. We present the NMR structure of KI-FHA from Arabidopsis KAPP at high resolution. Structure-based alignment suggests the possible folding nucleus of the FHA fold. The structure has been used in evolutionary trace (ET) analysis that has proven highly successful in predicting the recognition surfaces of proteins (20, 21), as well as dimerization sites (22). The phosphoprotein-binding surface of FHA domains is predicted by ET analysis and confirmed for KI-FHA by pThr peptide binding. ET analysis also suggests a second unexpected recognition surface on most FHA domains.
Materials and Methods
NMR of KI-FHA. A 134-residue KI-FHA construct of residues 180–313 of KAPP was expressed as a GST fusion. GST was removed by proteolysis and affinity chromatography (23). NMR samples of KI-FHA were 0.6 mM (the solubility limit at this pH) in 20 mM sodium phosphate (pH 6.3) with 120 mM NaCl and 7% D2O. 15N heteronuclear single quantum correlation (HSQC) spectra for hydrogen exchange protection and 13C HMQC-NOESY-HSQC were collected in 99% D2O. NMR experiments were performed at 22°C, except for the 25°C used in measuring J-couplings and residual dipolar couplings (RDCs). Chemical shift assignments of KI-FHA were reported (23). NMR spectra were collected with a Bruker DRX-500 spectrometer fitted with a Nalorac 8-mm 1H{13C/15N}-tuned probe with shielded z-gradient coil, or with a Varian INOVA 600 with 5-mm 1H{13C/15N}-tuned probe with shielded xyz-gradient coils. The 1,313 nontrivial nuclear Overhauser effect (NOE) restraints and 176 dihedral angle restraints were obtained as described in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
RDC Restraints. 1DNH, 1DCaHa, 1DCaC′, and 1DNC′ RDCs were measured by using 0.5 mM KI-FHA in the presence of Pf1 phage (24) at 9.6 mg/ml. Another set of 1DNH RDCs was measured with 0.4 mM KI-FHA in the presence of 5% (wt/vol) C12E6/hexanol (25). RDCs were obtained by comparing coupled spectra in the absence of orienting media against spectra in the presence of phage or polyethylene glycol (PEG)/hexanol media. The coupled spectra included 15N IPAP-HSQC, 13Ca-coupled HNCO, 15N-coupled HNCO, and 1Hα-coupled HNCA. Approximate values of the axial degree of alignment (Da) and rhombicity (R) of the alignment tensors were determined by the distribution of normalized dipolar couplings and a grid search (26). The final Da was 13 Hz for KI-FHA oriented with 5% (wt/vol) C12E6/hexanol. Da = –17 Hz for KI-FHA oriented with 9.6 mg/ml Pf1 phage. The optimized values of R were 0.50 and 0.60, respectively.
Structure Calculations. A torsion angle dynamics and simulated annealing (TAD/SA) protocol (27) was used to calculate structures by using cns 1.0 (28). Distances to degenerate protons were computed by using r–6 summation (29). The force constants during SA were: 50 kcal mol–1 A–2 for distances, 200 kcal mol–1 rad–2 for torsion angles, and 0.2 kcal mol–1 ppm–2 for secondary 13C /13α Cβ shifts. Final force constants for RDCs measured in phage, in units of kcal Hz–2, were: 0.3 for 1DNH, 0.2 for 1DCαHα, 2.5 for 1DNC′, and 1.5 for 1DCaC′. For 1DNH measured in 5% (wt/vol) C12E6/hexanol, the constant was 0.5 kcal Hz–2. These final force constants were adjusted such that rms deviations of the RDCs are not tighter than experimental errors. An independent floating alignment tensor was used for each class of RDCs during refinement using CNS; this procedure resulted in lower energy structures, improved convergence, and better satisfaction of RDC restraints. These independent tensors had reassuring, comparable magnitudes and orientations of the principal axes. To improve the convergence rate, the cooling steps were increased to 4,000 during TAD annealing, and to 9,000 during SA. Of paramount importance to increasing convergence rates, time steps during cooling stages were decreased to 5 fs in TAD annealing, and 2 fs in SA (30). Of the 100 calculated structures, 30 were selected based on the lowest total energy. molmol was used to inspect and display structures (31).
ET Analysis. A total of 301 sequences of FHA domains, spanning β-strands 3–10, and aligned at www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF00498, were combined with 12 Arabidopsis FHA sequences from the Munich Information Center for Protein Sequences (MIPS) Arabidopsis thaliana database (http://mips.gsf.de/proj/thal). To improve the multiple sequence alignment (MSA) using clustalw (www.ebi.ac.uk/clustalw), the most divergent FHA domains, such as those of kinesins, were removed. Truncated sequences that interfere with MSA were deleted (32). The MSA was submitted to the ET server (33) (www-cryst.bioc.cam.ac.uk/~jiye/evoltrace/evoltrace.html). The dendrogram was divided into 10 trace levels of sequence identity. Residues that are conserved within each subfamily, but vary among subfamilies, are considered class-specific residues at the chosen trace level (34).
KI-FHA Titration with pThr Peptide. A BAK1 (35) peptide fragment with sequence 307RLADGT(p)LVAVK317 that binds KI-FHA was synthesized and purified to 90% by reverse-phase HPLC. A series of 2D 15N-1H transverse relaxation-optimized spectroscopy (TROSY) spectra were collected at 22°C and 600 MHz with the molar ratio of protein to peptide titrated from 1:0 to 1:4. Resulting chemical shift changes were calculated as the radius, ΔωNH = (Δω H2 + (ΔωH/6)2)1/2, where ΔωH and ΔωN are the changes in ppm in the 1H and 15N dimensions.
Results and Discussion
Structure Determination and Refinement Using RDCs. A high-quality solution structure of KI-FHA was determined from 2,058 restraints featuring an average of 11 nontrivial NOEs per residue and 281 RDCs (Table 1). The RDCs enabled refinement of the backbone of the solution structural model to high accuracy. The distance restraints were derived from 15N-separated and 13C-separated NOESY spectra. Ambiguities among overlapped methyl NOEs were further resolved by the 13C dimensions of 3D 13C HMQC-NOESY-HSQC (36,) providing at least 100 more structurally significant, long-range NOEs. Weak alignment of the protein in the magnetic field is required for RDCs to be observed. RDCs were measured from samples weakly oriented by using either PEG/hexanol or Pf1 filamentous phage. Mobile regions experiencing structural averaging in nanoseconds or faster, as suggested by 15N{1H} NOE ratios of <0.65, were not refined with RDCs. A technical challenge of refining with numerous RDCs is that simulations yield fewer structures that satisfy all structural data. To boost the percentage of structures satisfying all restraints; i.e., to boost convergence, force constants and time steps used during the annealing portions of the TAD and SA stages of simulations were carefully adjusted. Decreasing the time steps, from the CNS default of 15 to 5 fs in the cooling phase of the TAD trajectory (30), improved the convergence rate dramatically from <10% to nearly 90%. Secondary structures including β4 were better defined when refined with RDCs. Approximately 83% of residues occupy most favored or favored regions of the Ramachandran plot (Table 1). The Q factor statistic on accuracy, using a scale from poorest of 1.0 to best of 0.0 (37), dramatically improved from refinement by using RDCs. For 1DNH RDCs from PEG/hexanol alignment, Q improved from 0.6 for the nonrefined structure to 0.15 for the RDC-refined structure, and from 0.55 to 0.11 for 1DNH RDCs from phage alignment.
Table 1. KI-FHA restraints and structural statistics.
| Restraints | Count | rmsd* |
|---|---|---|
| All distance restraints, Å | 1,367 | 0.03 ± 0.00 |
| Intra | 7 | |
| Sequential (|i - j| = 1) | 482 | |
| Medium (2 ≤ |i - j| ≤ 4) | 201 | |
| Long (|i - j| ≤ 4) | 623 | |
| Hydrogen bonds | 27·2 | |
| ϕ, ψ, χ1 dihedral angle, ° | 113, 47, 16 | 0.72 ± 0.12 |
| 13Cα chemical shift, ppm | 122 | 1.45 ± 0.08 |
| 13Cβ chemical shift, ppm | 112 | 1.54 ± 0.07 |
| Deviation from ideality | ||
| Bonds, Å | 0.003 ± 0.000 | |
| Angles, ° | 0.64 ± 0.03 | |
| Impropers, ° | 0.55 ± 0.04 | |
| All RDCs, Hz | 281 | |
| 1DNH (PEG-hexanol) | 62 | 2.01 ± 0.12 |
| 1DNH (phage) | 69 | 1.92 ± 0.13 |
| 1DCαHα (phage) | 55 | 2.26 ± 0.10 |
| 1DNC′ (phage) | 37 | 1.22 ± 0.12 |
| 1DCαC′ (phage) | 58 | 1.36 ± 0.07 |
| rmsd from mean structure, Å | ||
| All heavy atoms | 1.18 ± 0.18 | |
| Backbone atoms | 0.75 ± 0.18 | |
| β-Sheets†, all heavy atoms | 0.88 ± 0.11 | |
| β-Sheets†, backbone atoms | 0.38 ± 0.08 | |
| Ramachandran plot, %*‡ | ||
| Most favored regions | 67.8 | |
| Favored regions§ | 15.0 | |
| Additional allowed regions | 15.3 | |
| Generously allowed regions | 1.8 |
rmsd, rms deviation for the 30 accepted structures of 100 calculated. The five foreign, N-terminal linker residues and disordered residues 299-313 were excluded from the statistics.
Includes β-strand residues 181-186, 195-198, 208-211, 218-219, 230-235, 240-245, 253-254, 257-258, 273-274, 279-282, and 288-294.
The lowest 4% in quality occur in loops and termini not defined well by the NMR data.
The additionally allowed region from the program procheck was divided into the four gradations reported by webmol (46). The highest of these four is labeled as favored.
Description of the KI-FHA Structure. KI-FHA is a sandwich of a five-stranded mixed β-sheet (β-strands 3–6 and 9) with a six-stranded antiparallel β-sheet (β1, β2, β10, β11, β7, and β8) (Fig. 1). β1 and β11 are adjacent, bringing the N and C termini of the domain close together on the side opposite the phosphoprotein-binding surface. The hairpin of β1 and β2 (red in Fig. 1 a and b) crosses to β3 of the other sheet (blue). Short β4 runs parallel to β3, but all other strands run antiparallel to their neighbors. Residues essential for binding phosphorylated RLK kinase domains (9), namely Gly-211 and Arg-212 in the 3/4 loop, Ser-226 and His-229 in the 4/5 loop, and Asn-250 in the 6/7 loop, cluster on the edge of the five-stranded β-sheet in blue in Fig. 1b. Mostly conserved Asn-250 lies in a tight turn. The hairpin of β5 and β6 (blue in Fig. 1 a and b) crosses over into the hairpin of β7 and β8 at the edge of the other sheet (red). The 7/8 loop is a type I′ tight turn; i.e., with positive φ angles. Neighboring the 6/7 recognition loop, the 8/9 loop is not defined well, because of fewer NOE-detected contacts and greater mobility indicated by 15N relaxation (data not shown). β9 (blue) then connects through a type II turn to the hairpin of β10 and β11 in the middle of the six-stranded sheet in red in Fig. 1 a and b. C-terminal residues 299–313 appear to be unstructured (23), leaving open the possibility that the linker between KI-FHA and C-terminal PP2C domains might be flexible in KAPP. A flexible linker might allow freedom of orientation between KI-FHA and PP2C domains to facilitate KAPP interactions with RLKs and other components of transmembrane signaling complexes in plants, such as complexes containing CLV1, CLV2, CLV3, KAPP, and Rho GTPase (8).
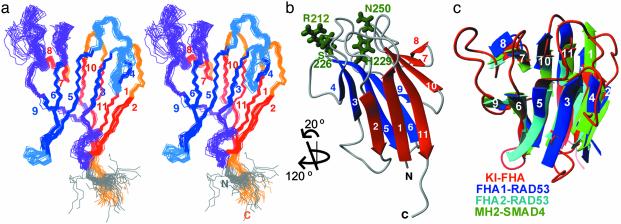
Fig. 1.
NMR structure of KI-FHA from Arabidopsis KAPP. (a) The stereoview shows a superposition of the 30 accepted KI-FHA models (PDB ID code 1MZK). Strands β1, β2, β7, β8, β10, and β11 of one β-sheet are red. Loops between the red strands and the C terminus are orange. Strands β3, β4, β5, β6, and β9 of the other β-sheet are blue. Loops between the blue strands are light blue. The 2/3, 6/7, and 8/9 loops crossing between the two β-sheets are violet. The five residues of the N-terminal linker are gray. (b) The ribbon plot of the backbone of representative model 1 of 1MZK is shown with the side chains of Arg-212, Ser-226, His-229, and Asn-250, which are essential for interaction with RLKs (9), colored green. (c) The β-sandwiches of Arabidopsis KI-FHA (red), the FHA1 (blue) and FHA2 (light blue) domains of yeast RAD53, and the MH2 domain (green) of the Smad4 tumor suppressor are superimposed. Loops are drawn only for KI-FHA.
Comparison of Fold with Other FHA Domains and MH2 of Smads. Although sequence identities are very low among distantly related FHA domains (Table 2), KI-FHA shares the fold of reported FHA structures (Fig. 1d). These FHA domains share not only the 11 β-strands, but also β-turns in the 3/4, 4/5, 6/7, 7/8, and 9/10 loops. Yeast RAD53 FHA1 and human Chk2 appear to be the closest of the reported structures to KI-FHA (Table 2). The comparatively high sequence identity of 19% between FHA1 of yeast RAD53 and KI-FHA of Arabidopsis KAPP accounts for the high Z score in the structural comparison (Table 2). Based on sequence, human CHFr seems more closely related to KI-FHA than does FHA2 of RAD53, although the segment swapping in CHFr obscures this fact (Table 2). KI-FHA also shares its fold with the MH2 domain of the tumor suppressor Smad4 (Table 2 and Fig. 1c), which was also identified as a pSer-binding module (38). This result suggests greater similarity in function than their negligible sequence identity would suggest. The MH2 domain of each Smad binds a phosphorylated type 1 TGFβ receptor with a surface that nearly coincides with that of the FHA domains (39). They share the 11-stranded β-sandwich that presents loops at the same edge of the sandwich to form a phosphoprotein-binding surface. Other parallels between FHA-dependent signaling and Smad/TGFβ receptor-dependent signaling are: (i) heterodimeric activation both of plant RLKs and of TGFβ receptors (35); and (ii) the presence of FHA and MH2 domains in transcription factors.
Table 2. Structural similarity of KI-FHA to other FHA domains and to MH2 of Smad4.
| Domain | Z score | rmsd*, Å | Identity percent† | No. of residues aligned† | Sequence identity with KI-FHA, % |
|---|---|---|---|---|---|
| FHA1-RAD53 | 11.3 | 2.2 | 23 | 100‡ | 19 |
| CHK2 | 11.4 | 2.2 | 19 | 116§ | 14 |
| FHA2-RAD53 | 8.7 | 2.3 | 17 | 103¶ | 14 |
| CHFr | 4.3 | 2.2 | 24 | 63∥ | ND** |
| MH2-Smad4 | 7.4 | 2.4 | 11 | 98†† | 9 |
Z score and rms deviation (rmsd) were obtained by a dali search (47). The PDB ID codes are 1MZK for KI-FHA, 1G6G-A for FHA1-RAD53, 1GCX for FHA from CHK2, 1QU5-A for FHA2-RAD53, 1LGP-A for CHFr, and 1YGS for MH2-Smad4.
Calculated from Cα-Cα distances.
Sequence identity among structurally aligned residues.
The aligned residues with KI-FHA residues in parentheses are 32(181)-40, 42(193)-51, 64(207)-79, 81(223)-95, 96(239)-104, 105(249)-117, 118(270)-133, and 139(286)-150.
The aligned residues with KI-FHA residues in parentheses are 92(181)-100, 101(192)-109, 110(206)-126, 136(223)-151, 155(239)-163, 164(249)-176, 177(270)-193, and 198(289)-206.
The aligned residues with KI-FHA residues in parentheses are 577(182)-583, 585(189)-596, 598(206)-631, 645(240)-655, 656(252)-660, 661(258)-665, 666(270)-682, 689(287)-696, and 698(295)-701.
The aligned residues with KI-FHA residues in parentheses are 14(181)-22, 31(196)-34, 36(207)-51, 53(223)-77, and 78(249)-86.
Not determined due to the segment swap.
The aligned residues with KI-FHA residues in parentheses are 322(181)-330, 334(193)-337, 339(197)-345, 346(207)-355, 361(218)-365, 380(223)-384, 385(229)-393, 394(239)-402, 403(249)-410, 426(258)-431, 432(280)-438, 499(288)-507, and 522(287)-531.
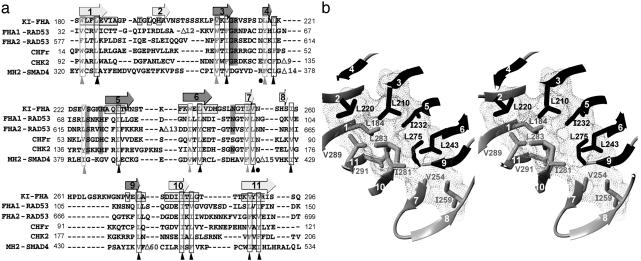
The fleeting homologies among FHA and Smad MH2 domains raises the question of what are minimal, shared determinants of their β-sandwich fold. The hydrophobic core and the folding nucleus of proteins is relatively well conserved in terms of hydrophobicity (40, 41). Consequently, we investigated preservation of hydrophobicity in this β-sandwich fold. Structure-based alignment (Fig. 2 and Table 2) reliably compares poorly homologous FHA and MH2 sequences. Nonfunctional core residues being conserved throughout diverse families, as demonstrated for the serpin, globin, and c cytochrome families, greatly increases the likelihood that such residues form the folding nucleus (40–42). FHA/MH2 fold members bind to a diverse range of proteins phosphorylated on Thr or Ser, implying diversification in function and structural details of phospho-binding surfaces. Consequently, the pSer-binding surface of MH2 domains bears no sequence resemblance to the pThr binding surface of FHA domains (Fig. 2a). Functional diversity also arises from binding of other proteins or domains to other nonconserved surfaces. For example, the Chk2 FHA domain binds proteins far from its FHA motif (18), and MH2 domains bind accessory domains and partners (43). Thus, the functional diversity is enough for conserved hydropathy to be structurally significant. Hydrophobicity is conserved at 20 residue positions of the β-sandwich (Fig. 2a). Eighteen positions are occupied by hydrophobic or aromatic residues and two positions by hydrophilic residues, all in or near β-strands. Most of the hydrophobic side chains pack to form the core (Fig. 2b). Twelve residues are most consistently hydrophobic among FHA domains and MH2 domains (black triangles in Fig. 2a). These 12 side chains, interdigitating within the β-sandwich (Fig. 2b), can be hypothesized to form the folding nucleus of FHA and MH2 domains. In KI-FHA, the hydrogen exchange protection of 10 of the 12 key hydrophobic residues corroborates this hypothesis (Fig. 2a). In the middle of this hydrophobic core, Ile-232, Leu-275, and Ile-281 of KAPP cluster and are characteristic of the FHA subfamily.
Fig. 2.
Proposed structural core of FHA domains and Smad MH2 domains. (a) Structure-based FHA sequence alignment marks locations of β-strands above. The most conserved residues and the invariant Gly have a gray background. KI-FHA residues with backbone amide protons protected from hydrogen exchange are marked with boxes. Residue positions where the side chains are almost exclusively hydrophobic are marked with black triangles. Hydrophobic positions where aromatic side chains with a polar group (W or Y) are common are marked with gray triangles. (b) In the same orientation as Fig. 3, the structural core of KI-FHA is shown in stereo. Side chains of residues with conserved hydrophobicity are shown as stick models. Contact surfaces are drawn with dots. The β-strands in black are from the five-stranded β-sheet. The β-strands in gray are from the six-stranded β-sheet.
Recognition Loops of KI-FHA. Structures of yeast RAD53 FHA1 (13, 16) and FHA2 (14), human CHFr (19), and human Chk2 (18) demonstrated that nominally conserved Gly, Arg, Ser, His, and Asn residues can either directly contact a bound phosphopeptide or support the structure of the binding surface. This finding appears to be true of KI-FHA as well. From the structure of RAD53 FHA1 (13) and structure-based alignment (Fig. 2a), His-229 NHδ1 of KI-FHA can be expected to form a hydrogen bond with the carbonyl oxygen of Ser-226, and may position conserved Ser-226 for hydrogen bonding of the pThr or pSer of an RLK binding partner. The NMR structure supports this hypothesis, because His-229 NHδ1 is oriented toward Ser-226. By analogy to the RAD53 FHA1 (13), the side chain of conserved Asn-250 of KI-FHA is close enough to form a hydrogen bond with the carbonyl oxygen of Glu-224 to position its side chain for interaction with the binding partner. Glu-224 of KI-FHA aligns with Arg-83 and Arg-617 of RAD53 (13) and Thr-138 in Chk2 (18) (Figs. 2a and 3 c and d), which are reported to interact with the pThr + 3 peptide residue that affects affinity. KAPP`s preference for serine at pThr + 3 (11) might be fostered by hydrogen bonds with Glu-224 or with Thr-285 of the β10/11 loop.
Fig. 3.
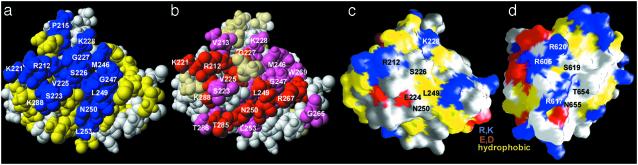
Phosphoprotein-binding surface of KI-FHA (a, b, and c) and of FHA2 of RAD53 (d). (a) The class-specific residues at trace level 9 are blue. (Fig. 6 summarizes the alignment of class consensus residues at trace level 9.) The residues identical among KAPP of maize, rice, and Arabidopsis are yellow in a.(b) KI-FHA residues with amide NMR peaks most shifted by saturating amounts of the pThr peptide from BAK1 (2 mM peptide:0.5 mM KI-FHA); i.e., with ΔωHN > 0.16 ppm are red. Those residues with 0.16 ppm > ΔωHN > 0.06 ppm are pink. Surface properties of KI-FHA (c) and RAD53 FHA2 (d) are yellow for hydrophobic, blue for positively charged, and red for negatively charged residues.
ET Analysis of Phosphoprotein-Binding Surface. FHA domain surfaces important in recognition and specificity were mapped by ET analysis (34). ET analysis relies on two principles. First, recognition surfaces are more conserved than other surfaces. Second, a protein family can be divided into functional classes based on sequence, because differences in binding specificity correlate with systematic variations in sequence at binding sites (20, 34). The dendrogram for a MSA is divided into classes, with the branching determined by the trace level of sequence identity. Class consensus residues are those conserved within a class. Class-specific residues lie at positions where a class consensus residue is present in all classes but differs in one or more classes (34). When mapped on a representative structure, class-specific and conserved residues cluster at binding sites (34).
Class-specific residues may confer distinctive specificities to FHA family members by altering surface properties, such as those of the phospho-recognition loops (see Fig. 3 c and d). ET analysis used an alignment of 209 sequences of FHA domains that excludes divergent kinesins. The dendrogram and trace levels are shown in Fig. 5, which is published as supporting information on the PNAS web site. Because of the great sequence diversity among FHA domains, clusters of class-specific residues did not emerge until the ninth highest of 10 trace levels, i.e., at sequence identities of >84% within each class (see Figs. 5–7, which are published as supporting information on the PNAS web site). Many class-specific FHA residues at trace level 9 lie at or near the phosphoprotein-binding surface of KI-FHA, i.e., Arg-212, Ser-214–Ala-219, Lys-221–Ser-223, Val-225–His-229, and Met-246–Leu-253 (blue in Fig. 3a). Conserved residues (10) essential to binding of phosphorylated RLKs (9), i.e., Arg-212, Ser-226, His-229, and Asn-250 of KAPP, are in fact class specific (Fig. 3a). Gly-211 is the only absolutely conserved residue (Figs. 2a and 3a). Residues flanking these residues were shown in other FHA domains to contact phosphopeptide ligands and are seen to be class specific. Counterparts to class-specific Leu-249 of KAPP (Figs. 2 and 3a) are Thr-654 of RAD53 FHA2 (14), Thr-106 of RAD53 FHA1 (13), and Gly-165 in Chk2 (18) (Fig. 2a) that interact with the key pThr + 3 position of peptides. Class-specific Gly-227 of KAPP (Fig. 3a) corresponds to Asn-86 and Arg-620 of RAD53 (Fig. 2a) that can contact pThr peptides (13, 17). Class-specific Ser-223 of KI-FHA (Fig. 3a) may contact a pThr + 1 residue, by analogy with RAD53 FHA1 (13) and FHA2 (17). The identity of Ser-223 of KI-FHA with Ser-82 of RAD53 FHA1 accounts for their very similar preference for pThr + 1 residues (13).
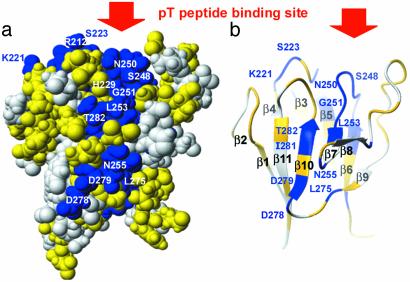
pThr Peptide-Binding Surface. We tested the hypothesis that residues that are class specific among FHA domains (blue in Fig. 3a), or conserved in KAPP of plants (yellow in Fig. 3a), interact with RLK partners. A pThr peptide sequence from the N-terminus of the kinase domain of BAK1; i.e., 307RLADGT-(p)LVAVK317, was found by titration calorimetry to bind KI-FHA. Titration of 15N-enriched KI-FHA with this pThr peptide shifted the amide peak positions of loops of this face of the β-sandwich (see Fig. 3b, and quantified in Fig. 8, which is published as supporting information on the PNAS web site). Residues with perturbed amide chemical shifts include: Gly-211–Val-213 of the 3/4 loop; Lys-221–Ser-223, Val-225, and Gly-227–His-229 of the 4/5 loop; Met-246–Leu-253 of the 6/7 loop; Asp-263, Gly-265, Arg-267, and Trp-269 of the 8/9 loop; and Leu-283–Thr-287 of the 10/11 loop (see Figs. 3b and 8). Participation of the 3/4, 4/5, 6/7, and sometimes 10/11 loops in phosphopeptide binding has been reported in FHA studies. The involvement of the 8/9 loop seems unique to KAPP of plants, and is facilitated by the greater length and exposure of this loop in KAPP (Fig. 1a; compare lower right of Fig. 3 c with d). The class-specific residues in blue and KAPP-conserved residues in yellow in Fig. 3a correspond very well with the pT peptide-perturbed surface (red and pink in Fig. 3b). Of the 27 pThr peptide-perturbed residues, 16 are class specific, and the other 11 are conserved in KAPP. This result corroborates the hypothesis of a broad RLK-binding surface.
Other Recognition Surfaces Suggested by ET Analysis. An unexpected potential functional surface on most FHA domains (Fig. 4 a and b), oriented at a 90° angle from the phosphoprotein-binding surface, is suggested by the patch of class-specific residues on the surface around β7 (Gly-251, Leu-253, and Asn-255 in KI-FHA) and adjacent in β10 (Asp-278, Asp-279, and Thr-282 in KI-FHA) (Fig. 4). It is conceivable that a variety of FHA domains could have the capacity to use this second surface for self-assembly. One possible mode of self-assembly is hydrogen-bonded pairing with another β-strand of the same chain, like the Smad activation domain (SAD) pairs with β8 of the MH2 domain in Smad4 (44). In KAPP, a sequence of similar size and location as SAD in Smads is predicted to contain a β-strand. Perhaps this KAPP sequence might likewise be able to pair with β8 of KI-FHA. Another possibility is oligomerization as reported for FHA-containing CHK2 (45), CHFr (19), and MH2 of Smads (38). Contrasting with the β7/β8 hairpin of monomeric FHA structures, the crystalline FHA domain of human CHFr dimerizes with a single long β-strand that inserts between the β7/β8 strand and β-strand 10 from the other chain of the dimer (19). The β7/β8 segment and β10 of FHA domains coincides with the second class-specific surface.
Fig. 4.
Unexpected class-specific surface suggests another interaction surface. This view is rotated 90° about the x axis relative to Fig. 3, placing the six-stranded β-sheet in the foreground. The color code is the same as that of Fig. 3a. (a) The CPK surface. (b) A backbone ribbon of KI-FHA in the same orientation as a.
Distinct from the site of Fig. 4a is the site of BRCA1 binding reported on the other (five-stranded) β-sheet of the FHA domain of human Chk2 (18). This latter site in Chk2 was found far from the phosphopeptide-binding site through the I157T mutation of Li–Fraumeni cancer patients that disrupts binding of BRCA1, CDC25A, and p53 (18). Extended ET analysis results at trace level 7 suggest that Ile-157 of Chk2, corresponding to Lys-240 in KI-FHA of KAPP (Fig. 2a), plus neighboring residues, might form another binding site in a majority of FHA domains (Fig. 7). Lys-240 and Asn-235 of the 5/6 loop of KI-FHA cluster on a ridge at the center of this patch that could extend to Pro-272, Glu-274 (of β9), or Thr-233 of KI-FHA.
Implications for RLK Signaling in Plants. The ability of KAPP to interact in vivo with several different RLKs in Arabidopsis, including HAESA (2), CLAVATA1 (8), FLS2 (6), WAK1 (5) and SERK1 (7), indicates that KAPP functions in multiple signaling pathways in plants. KAPP probably requires more than pSer or pThr residues on the RLKs for binding, because it interacts with just a subset of the kinases tested (3). KI-FHA of KAPP presents a broad surface for RLK recognition (Fig. 3 a–c). Plant KAPP KI-FHA sequences are closely related. Among KAPPs of plants, clearly the greatest conservation of surface residues on KI-FHA occurs on the face that binds phosphorylated RLKs (Fig. 3a), and the second face encompassing a patch of class-specific residues (Fig. 4), followed by the third class-specific patch of FHA domains (Fig. 7). This finding suggests the importance of these surfaces in assembly of KAPP into RLK signaling supercomplexes in plant membranes.
Supplementary Material
Acknowledgments
We thank Jia Li for assistance in expressing KI-FHA, and the National Magnetic Resonance Facility at Madison (Madison, WI) for access to the cryoprobe-equipped Bruker DMX-500. This work was supported by National Science Foundation Grants MCB0111589 (to S.R.V.D.) and MCB98098848 and MCB0112278 (to J.C.W.). The Varian Inova 600 and Bruker DRX-500 spectrometers were funded in part by National Science Foundation Grants DBI0070359 and CHE8908304. The National Magnetic Resonance Facility at Madison receives support from the National Center for Research Resources, the National Science Foundation, and the U.S. Department of Agriculture.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FHA, forkhead-associated; KAPP, kinase-associated protein phosphatase; KI-FHA, kinase-interacting FHA domain of KAPP; ET, evolutionary trace; NOE, nuclear Overhauser effect; RLK, receptor-like protein kinase; RDC, residual dipolar coupling; SA, simulated annealing; TAD, torsion angle dynamics; HSQC, heteronuclear single quantum correlation; pThr, phosphothreonine.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1MZK).
References
- 1.Becraft, P. W. (2002) Annu. Rev. Cell Dev. Biol. 18, 163–192. [DOI] [PubMed] [Google Scholar]
- 2.Stone, J. M., Colinge, M. A., Smith, R. D., Horn, M. A. & Walker, J. C. (1994) Science 266, 793–795. [DOI] [PubMed] [Google Scholar]
- 3.Braun, D. M., Stone, J. M. & Walker, J. C. (1997) Plant J. 12, 83–95. [DOI] [PubMed] [Google Scholar]
- 4.Williams, R. W., Wilson, J. M. & Meyerowitz, E. M. (1997) Proc. Natl. Acad. Sci. USA 94, 10467–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park, A. R., Cho, S. K., Yun, U. J., Jin, M. Y., Lee, S. H., Sachetto-Martins, G. & Park, O. K. (2001) J. Biol. Chem. 276, 26688–26693. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Gómez, L., Bauer, Z. & Boller, T. (2001) Plant Cell 13, 1155–1163. [PMC free article] [PubMed] [Google Scholar]
- 7.Shah, K., Russinova, E., Gadella, T. W., Jr., Willemse, J. & De Vries, S. C. (2002) Genes Dev. 16, 1707–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trotochaud, A. E., Hao, T., Wu, G., Yang, Z. & Clark, S. E. (1999) Plant Cell 11, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, J., Smith, G. P. & Walker, J. C. (1999) Proc. Natl. Acad. Sci. USA 96, 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofman, K. & Bucher, P. (1995) Trends Biochem. Sci. 20, 347–349. [DOI] [PubMed] [Google Scholar]
- 11.Durocher, D., Henckel, J., Fersht, A. R. & Jackson, S. P. (1999) Mol. Cell 4, 387–394. [DOI] [PubMed] [Google Scholar]
- 12.Sun, Z., Hsiao, J., Fay, D. S. & Stern, D. F. (1998) Science 281, 272–274. [DOI] [PubMed] [Google Scholar]
- 13.Durocher, D., Taylor, I. A., Sarbassova, D., Haire, L. F., Westcott, S. L., Jackson, S. P., Smerdon, S. J. & Yaffe, M. B. (2000) Mol. Cell 6, 1169–1182. [DOI] [PubMed] [Google Scholar]
- 14.Liao, H., Byeon, I. J. & Tsai, M. D. (1999) J. Mol. Biol. 294, 1041–1049. [DOI] [PubMed] [Google Scholar]
- 15.Durocher, D. & Jackson, S. P. (2002) FEBS Lett. 513, 58–66. [DOI] [PubMed] [Google Scholar]
- 16.Liao, H., Yuan, C., Su, M. I., Yongkiettrakul, S., Qin, D., Li, H., Byeon, I. J., Pei, D. & Tsai, M. D. (2000) J. Mol. Biol. 304, 941–951. [DOI] [PubMed] [Google Scholar]
- 17.Byeon, I. J., Yongkiettrakul, S. & Tsai, M. D. (2001) J. Mol. Biol. 314, 577–588. [DOI] [PubMed] [Google Scholar]
- 18.Li, J., Williams, B. L., Haire, L. F., Goldberg, M., Wilker, E., Durocher, D., Yaffe, M. B., Jackson, S. P. & Smerdon, S. J. (2002) Mol. Cell 9, 1045–1054. [DOI] [PubMed] [Google Scholar]
- 19.Stavridi, E. S., Huyen, Y., Loreto, I. R., Scolnick, D. M., Halazonetis, T. D., Pavletich, N. P. & Jeffrey, P. D. (2002) Structure (London) 10, 891–899. [DOI] [PubMed] [Google Scholar]
- 20.Sowa, M. E., He, W., Slep, K. C., Kercher, M. A., Lichtarge, O. & Wensel, T. G. (2001) Nat. Struct. Biol. 8, 234–237. [DOI] [PubMed] [Google Scholar]
- 21.Aloy, P., Querol, E., Aviles, F. X. & Sternberg, M. J. (2001) J. Mol. Biol. 311, 395–408. [DOI] [PubMed] [Google Scholar]
- 22.Dean, M. K., Higgs, C., Smith, R. E., Bywater, R. P., Snell, C. R., Scott, P. D., Upton, G. J., Howe, T. J. & Reynolds, C. A. (2001) J. Med. Chem. 44, 4595–4614. [DOI] [PubMed] [Google Scholar]
- 23.Lee, G., Li, J., Walker, J. C. & Van Doren, S. R. (2003) J. Biomol. NMR 25, 253–254. [DOI] [PubMed] [Google Scholar]
- 24.Hansen, M. R., Hanson, P. & Pardi, A. (2000) Methods Enzymol. 317, 220–240. [DOI] [PubMed] [Google Scholar]
- 25.Rückert, M. & Otting, G. (2000) J. Am. Chem. Soc. 122, 7793–7797. [Google Scholar]
- 26.Clore, G. M., Gronenborn, A. M. & Bax, A. (1998) J. Magn. Reson. 133, 216–221. [DOI] [PubMed] [Google Scholar]
- 27.Stein, E. G., Rice, L. M. & Brunger, A. T. (1997) J. Magn. Reson. 124, 154–164. [DOI] [PubMed] [Google Scholar]
- 28.Brünger, A. T., Adams, P. D., Clore, G. M., Delano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 29.Fletcher, C. M., Jones, D. N. M., Diamond, R. & Neuhaus, D. (1996) J. Biomol. NMR 8, 292–310. [DOI] [PubMed] [Google Scholar]
- 30.Choy, W. Y., Tollinger, M., Mueller, G. A. & Kay, L. E. (2001) J. Biomol. NMR 21, 31–40. [DOI] [PubMed] [Google Scholar]
- 31.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]
- 32.Madabushi, S., Yao, H., Marsh, M., Kristensen, D. M., Philippi, A., Sowa, M. E. & Lichtarge, O. (2002) J. Mol. Biol. 316, 139–154. [DOI] [PubMed] [Google Scholar]
- 33.Innis, C. A., Shi, J. & Blundell, T. L. (2000) Protein Eng. 13, 839–847. [DOI] [PubMed] [Google Scholar]
- 34.Lichtarge, O., Bourne, H. R. & Cohen, F. E. (1996) J. Mol. Biol. 257, 342–358. [DOI] [PubMed] [Google Scholar]
- 35.Li, J., Wen, J. Q., Lease, K. A., Doke, J. T., Tax, F. E. & Walker, J. C. (2002) Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- 36.Morshauser, R. C., Hu, W., Wang, H., Pang, Y., Flynn, G. C. & Zuiderweg, E. R. (1999) J. Mol. Biol. 289, 1387–1403. [DOI] [PubMed] [Google Scholar]
- 37.Cornilescu, G., Marquardt, J. L., Ottiger, M. & Bax, A. (1998) J. Am. Chem. Soc. 120, 6836–6837. [Google Scholar]
- 38.Wu, J. W., Hu, M., Chai, J., Seoane, J., Huse, M., Li, C., Rigotti, D. J., Kyin, S., Muir, T. W., Fairman, R., et al. (2001) Mol. Cell 8, 1277–1289. [DOI] [PubMed] [Google Scholar]
- 39.Huse, M., Muir, T. W., Xu, L., Chen, Y. G., Kuriyan, J. & Massague, J. (2001) Mol. Cell 8, 671–682. [DOI] [PubMed] [Google Scholar]
- 40.Shakhnovich, E., Abkevich, V. & Ptitsyn, O. (1996) Nature 379, 96–98. [DOI] [PubMed] [Google Scholar]
- 41.Mirny, L. & Shakhnovich, E. (2001) J. Mol. Biol. 308, 123–129. [DOI] [PubMed] [Google Scholar]
- 42.Ptitsyn, O. B. & Ting, K. L. (1999) J. Mol. Biol. 291, 671–682. [DOI] [PubMed] [Google Scholar]
- 43.Shi, Y. (2001) BioEssays 23, 223–232. [DOI] [PubMed] [Google Scholar]
- 44.Qin, B., Lam, S. S. & Lin, K. (1999) Structure Fold Des. 7, 1493–1503. [DOI] [PubMed] [Google Scholar]
- 45.Xu, X., Tsvetkov, L. M. & Stern, D. F. (2002) Mol. Cell. Biol. 22, 4419–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walther, D. & Cohen, F. E. (1999) Acta Crystallogr. D 55, 506–517. [DOI] [PubMed] [Google Scholar]
- 47.Holm, L. & Sander, C. (1995) Trends Biochem. Sci. 20, 478–480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.