Abstract
Forkhead transcription factor FKHR (Foxo1) is a key regulator of glucose homeostasis, cell-cycle progression, and apoptosis. It has been shown that FKHR is phosphorylated via insulin or growth factor signaling cascades, resulting in its cytoplasmic retention and the repression of target gene expression. Here, we investigate the fate of FKHR after cells are stimulated by insulin. We show that insulin treatment decreases endogenous FKHR proteins in HepG2 cells, which is inhibited by proteasome inhibitors. FKHR is ubiquitinated in vivo and in vitro, and insulin enhances the ubiquitination in the cells. In addition, the signal to FKHR degradation from insulin is mediated by the phosphatidylinositol 3-kinase pathway, and the mutation of FKHR at the serine or threonine residues phosphorylated by protein kinase B, a downstream target of phosphatidylinositol 3-kinase, inhibits the ubiquitination in vivo and in vitro. Finally, efficient ubiquitination of FKHR requires both phosphorylation and cytoplasmic retention in the cells. These results demonstrate that the insulin-induced phosphorylation of FKHR leads to the multistep negative regulation, not only by the nuclear exclusion but also the ubiquitination-mediated degradation.
Protein kinase B (PKB, also called Akt) plays a critical role in mediating various effects of insulin and related growth factors, downstream from phosphatidylinositol 3-kinase (PI3K) (1, 2). PKB promotes cell survival and proliferation and regulates metabolic homeostasis in part by modulating transcriptional activity of the FOXO subfamily of Forkhead transcription factors, including FKHR (Foxo1), FKHRL1 (Foxo3a), and AFX (Foxo4), through phosphorylation (3, 4). In the absence of phosphorylation, these FOXO transcription factors localize in the nucleus and interact with the insulin response sequences within the promoters of multiple target genes. Once bound to the target gene promoters via insulin response sequences, the FOXOs act as a potent activator of transcription. On the other hand, when PKB is activated by insulin, the FOXO proteins are directly phosphorylated, resulting in their nuclear export and cytoplasmic retention through the binding to 14-3-3 proteins and the inhibition of target gene expression (5–9).
Recent studies have revealed that FOXOs are implicated in modulating various cellular activities in different cell types. For instance, in hepatocytes, FOXOs regulate the transcription of gluconeogenic key factors, including glucose-6-phosphatase, phosphoenolpyruvate carboxykinase, and peroxisome proliferator-activated receptor-γ coactivator-1 (10–13). Nakae et al. (14) have also reported recently that FKHR (Foxo1) controls hepatic glucose production and β cell compensation for insulin resistance in type 2 diabetes by the genetic analysis with the gain- and loss-of-functions of FKHR alleles in mice. In Caenorhabditis elegans, several studies have provided genetic evidence that FOXO homologue Daf-16 acts as a determinant of longevity, probably by inducing several stress-resistance genes (15–17). More recent studies demonstrated that FOXOs regulate the gene expression of catalase and manganese superoxide dismutase, which protect cells against oxidative stress (18, 19), and suggested that FOXOs are able to control lifespan in mammals. In addition, it has been shown that FOXOs can mediate the transcriptional activity of nuclear hormone receptors by serving as either a coactivator or a corepressor (20–22).
The relevance of the FOXO family to diverse physiological events in the transcription-based function is of significant interest. In postphosphorylation, however, the fate of FOXO proteins remains to be explored. Here, we show that FKHR (Foxo1) is a substrate of insulin-dependent polyubiquitination and degradation. Significantly, FKHR 3A mutant, resistant to PKB phosphorylation, was unable to be polyubiquitinated in vivo and in vitro, suggesting the requirement of the “phosphorylated” state for ubiquitination. Furthermore, the cytoplasmic retention in addition to phosphorylation achieves the effective ubiquitination of FKHR. Taken together, our results provide evidence for the mechanism whereby insulin signaling leads to FKHR polyubiquitination, resulting in down-regulation of the protein level via proteasome-dependent degradation.
Materials and Methods
Cell Culture and Western Blot Analysis. HepG2 and HEK293T cells were cultured in DMEM supplemented with 10% FBS. Before the insulin treatment of HepG2 cells, medium was replaced with serum-free DMEM containing 0.1% BSA and incubated for 12 h, and then added with insulin (100 nM) with or without proteasome inhibitors (10 μM MG132, 10 μM lactacystin, or 10 μM epoxomicin). In some experiments, cells were pretreated with LY294002 (20 μM) or wortmannin (100 nM) for 30 min before insulin stimulation. After washing with PBS, cells were lysed with lysis buffer (50 mM Tris·HCl, pH 7.5/150 mM NaCl/1% Triton X-100/0.1% SDS/0.5% sodium deoxycholate/10% glycerol/5 mM NaF/1 mM sodium orthovanadate and protease inhibitors). Forty micrograms of whole cell extracts were resolved by SDS/PAGE, followed by electrotransfer onto poly(vinylidene difluoride) membranes and probing with an anti-FKHR antibody. Chemiluminescent detection relied on horseradish peroxidase-conjugated secondary antibodies.
Antibodies. A rabbit polyclonal antibody specific for mouse FKHR was raised against the bacterially expressed GST-FKHR (541–652 aa) fusion protein as described (13). Anti-FLAG (M2) and anti-β-actin (AC-74) were purchased from Sigma. Anti-hemagglutinin (anti-HA) (3F10) was obtained from Roche Molecular Biochemicals. Anti-phospho-PKB/Akt (Ser-473), anti-PKB/Akt, and anti-phospho-FKHR (Ser-256) were from Cell Signaling Technology (Beverly, MA). Anti-Xpress was from Invitrogen.
Cloning and Plasmids. pcDNA3-FLAG-FKHR was generated by RT-PCR-based cloning of mouse FKHR cDNA into pcDNA3-FLAG vector as described (23). The putative PKB phosphorylation sites at Thr-24, Ser-253, and Ser-316 of FKHR were mutated to alanines by PCR mutagenesis. The critical residues in nuclear localization signal (NLS) or nuclear export signal (NES) of FKHR were also mutated to alanines by PCR mutagenesis. pcDNA3-HA-ubiquitin was described (24). GST-HA-ubiquitin was made by PCR-based subcloning into pGEX-4T vector (Amersham Pharmacia). cDNA of Δp85, which is p85 subunit of PI3K lacking binding site for its catalytic p110 subunit (25), was kindly provided by W. Ogawa (Kobe University, Kobe, Japan) and was subcloned into pcDNA3.1-HisC vector (Invitrogen) to express Xpress epitope-tagged Δp85 (Xp-Δp85).
Immunoprecipitation. To detect the ubiquitination of FKHR in vivo, HepG2 cells were transfected with FLAG-FKHR and HA-ubiquitin expression plasmids by using FuGENE 6 reagents (Roche Molecular Biochemicals). Thirty-six hours after transfection, cells were treated with MG132, lactacystin, or epoxomicin for 12 h, then lysed in lysis buffer. Where indicated, before treatment with insulin and MG132, cells were serum-starved for 12 h. Whole cell extracts were subjected to immunoprecipitation with anti-FLAG antibody. To test whether FKHR is covalently linked to ubiquitin, immunoprecipitates were eluted from the beads by heating in the presence of 1% SDS and reimmunoprecipitated with the same antibody as described (26). All of the immune complexes were resolved by SDS/PAGE, followed by Western blot analysis.
In Vitro Ubiquitination Assay. The in vitro ubiquitination assay was performed as described (27) with minor modifications. HEK293T cells were transfected with FLAG-FKHR wild-type or T24A/S253A/S316A (3A) mutant as described above. Forty-eight hours later, cells were lysed in lysis buffer, and whole cell extracts were subjected to immunoprecipitation with anti-FLAG antibody. The immune complexes were washed with lysis buffer, followed with reaction buffer. The reaction mixture contained the immunoprecipitates, 50 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 0.5 mM DTT, ATP regenerating system (2 mM ATP, 10 mM creatine phosphate, 10 units/ml creatine kinase, and 1 unit/ml inorganic pyrophosphatase), 20 μM MG132, and 33% (vol/vol) crude rabbit reticulocyte lysate (RRL, Promega), with 6 μg of bacterially expressed GST, GST-HA-ubiquitin, or 3 μg of ubiquitin (Sigma). After incubation at 30°C for 1 h, the beads were extensively washed with reaction buffer containing 1% Triton X-100. In some cases, the reaction products were eluted with FLAG peptide (Sigma), and ubiquitinated FKHR was purified with glutathione-Sepharose beads (Amersham Pharmacia) or an anti-HA antibody. All of the reaction products were analyzed by Western blotting.
Immunofluorescence. HepG2 cells were plated onto glass coverslips and transfected by using FuGENE 6 reagents. Forty-eight hours after transfection, cells were fixed with 3.7% paraformaldehyde in PBS for 20 min and permeabilized by treatment with 0.1% Triton X-100 in PBS for 30 min. After blocking with 3% BSA, 0.1% Tween 20 in PBS for 1 h, cells were incubated with anti-FLAG antibody, followed by staining with Alexa Fluor 488 anti-mouse secondary antibody (Molecular Probes).
Results
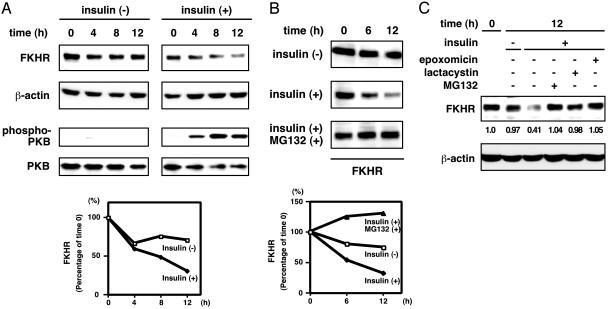
Down-Regulation of FKHR Expression by Insulin. To explore the fate of FKHR subsequent to phosphorylation and cytoplasmic retention in response to insulin, we first examined the kinetics of FKHR stability after the stimulation. Serum-starved HepG2 cells were cultured in the presence or absence of insulin, and cell extracts were harvested at different time points after the stimulation to monitor the amount of endogenous FKHR by using anti-FKHR antibody. Interestingly, although little change in stability was observed in the untreated cells, insulin stimulation decreased the level of FKHR protein in a time-dependent manner (Fig. 1A). At the same time, PKB was activated by insulin stimulation.
Fig. 1.
Insulin induces FKHR turnover through proteasomal degradation. (A and B) HepG2 cells were serum-starved for 12 h and treated with 100 nM insulin (A), with or without a proteasome inhibitor MG132 (B), for the indicated periods. Whole cell extracts were analyzed by immunoblotting. (C) HepG2 cells were serum-starved for 12 h and treated with insulin in the presence or absence of indicated proteasome inhibitors for 12 h. Whole cell extracts were analyzed by immunoblotting.
Because it is known that various cellular proteins, including transcription factors, are tightly regulated by proteolysis through proteasome in response to external signaling molecules (28, 29), we next sought to determine whether a proteasome-mediated degradation is involved in the insulin-dependent decline of FKHR level. To address this question, serum-starved HepG2 cells were cultured with insulin in the presence or absence of a proteasome inhibitor, MG132. As shown in Fig. 1B, the proteasome inhibition resulted in the drastic stabilization of FKHR despite insulin stimulation. Other proteasome-specific inhibitors, lactacystin and epoxomicin, also stabilized FKHR protein (Fig. 1C). This finding provided an indication that FKHR is regulated through proteasome-mediated degradation, which is triggered by insulin stimulation.
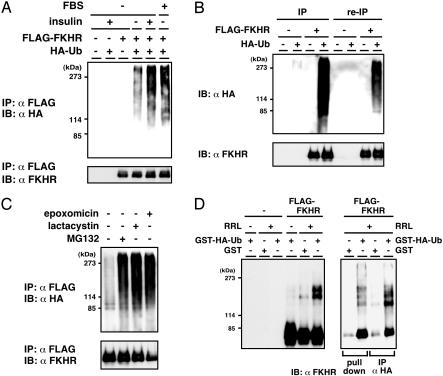
Ubiquitination of FKHR in Vivo and in Vitro. Because the 26S proteasome recognizes and degrades proteins that are conjugated with polyubiquitin chains (30), we investigated whether FKHR is ubiquitinated in vivo. HepG2 cells were cotransfected with FLAG-tagged FKHR and HA-tagged ubiquitin expression plasmids and serum-starved before insulin and MG132 treatment. FLAG-FKHR was immunoprecipitated from whole cell extracts and subjected to immunoblotting using anti-HA antibody to detect ubiquitin-conjugated FKHR. A ladder of high molecular weight ubiquitinated products was detected when cells were treated with insulin, whereas only slight signal was seen at an immunoprecipitate from untreated cells (Fig. 2A), indicating the insulin-dependent polyubiquitination of FKHR. A similar effect to insulin was observed by the addition of FBS (see Fig. 2 A). Furthermore, to exclude the possibility that other ubiquitinated proteins coimmunoprecipitated with FKHR may be detected, reimmunoprecipitation assay was performed. After the immunoprecipitated complex was boiled in a buffer containing 1% SDS to disrupt all protein–protein interaction, FKHR was immunoprecipitated again and tested for the ubiquitin conjugation. The shifted ladder larger than unmodified FKHR was detected in the reimmunoprecipitation complex, suggesting that FKHR is indeed a target of the ubiquitination in HepG2 cells (Fig. 2B). Several proteasome-specific inhibitors were used to confirm that the ubiquitinated FKHR could be degraded by the 26S proteasome. Consistent with MG132 treatment, lactacystin and epoxomicin, but not solvent DMSO, induced the accumulation of ubiquitinated FKHR (Fig. 2C), suggesting that ubiquitinated FKHR is a substrate of the 26S proteasome in vivo.
Fig. 2.
FKHR is ubiquitinated in vivo and in vitro. (A) HepG2 cells were transfected with the FLAG-FKHR and HA-ubiquitin expression plasmids. Twenty-four hours after transfection, cells were serum-starved for 12 h and incubated with MG132 with or without insulin or FBS for a further 12 h. Whole cell extracts were subjected to anti-FLAG immunoprecipitation (IP) and followed by anti-HA (Upper) or anti-FLAG (Lower) immunoblotting. (B) HepG2 cells were transfected with the FLAG-FKHR and HA-ubiquitin expression plasmids and treated with MG132 for 10 h before cell lysis. Whole cell extracts were subjected to anti-FLAG immunoprecipitation. The immunoprecipitated materials were eluted from the beads by heating in the presence of 1% SDS and reimmunoprecipitated (re-IP) with the same antibody. All of the immune complexes were analyzed as in A. (C) HepG2 cells transfected with the FLAG-FKHR and HA-ubiquitin were treated with indicated proteasome inhibitors, and ubiquitination was detected as in A.(D) Cell extracts from HEK293T cells transfected with the FLAG-FKHR plasmid were subjected to anti-FLAG immunoprecipitation. The immune complexes were incubated at 30°C for 1 h with or without RRL and either GST or GST-HA-ubiquitin. (Right) The reaction products were eluted by FLAG peptide, again subjected to pull-down by glutathione-Sepharose or to immunoprecipitation by using anti-HA antibody. All of the reaction products were analyzed by immunoblotting with anti-FKHR antibody.
Next, to confirm whether ubiquitin chain is directly attached to FKHR molecule, we performed an in vitro cell-free assay. FLAG-FKHR was immunoprecipitated from transfected HEK293T cells and incubated with RRL in the presence of ATP regenerating system and bacterially expressed GST-HA-ubiquitin. By immunoblotting with anti-FKHR antibody, the bands of large molecular size were detected (Fig. 2D Left), indicating that FKHR was modified, probably by ubiquitination. Furthermore, to verify that FKHR was indeed ubiquitinated in this assay, the reaction products were eluted by FLAG peptide, followed by pull-down with glutathione-Sepharose or by immunoprecipitation with anti-HA antibody. As shown in Fig. 2D Right, the bands of high molecular weight proteins were also probed with anti-FKHR antibody, clearly demonstrating that the modified molecules were ubiquitin-conjugated FKHR.
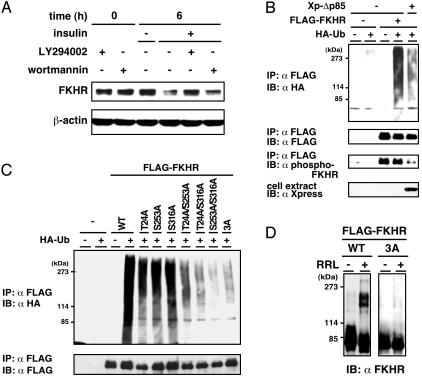
Phosphorylation-Dependent Ubiquitination and Degradation of FKHR. Our initial finding of a strong correlation between insulin signaling and FKHR instability prompted us to investigate whether the PI3K-PKB pathway is involved in the signaling for FKHR degradation. To this end, HepG2 cells were preincubated with PI3K-specific inhibitors, LY294002 or wortmannin, followed by treatment with insulin. Whereas FKHR level declined by insulin stimulation as presented above, PI3K inhibitors substantially inhibited the effect of insulin on FKHR degradation (Fig. 3A). Next, to examine the involvement of PI3K in FKHR ubiquitination signal, a dominant negative mutant of PI3K (Xp-Δp85) was cotransfected with FLAG-FKHR and HA-ubiquitin in HepG2 cells. Coexpression of Δp85 decreased FKHR ubiquitination accompanied with the reduced phosphorylation (Fig. 3B). These results suggest that insulin stimulation accelerates FKHR ubiquitination and degradation via a PI3K-signaling pathway. Moreover, to examine whether insulin-dependent ubiquitination of FKHR depends on the site-specific phosphorylation by PKB, an in vivo ubiquitination assay was performed by using FKHR phosphorylation site mutants. The individual T24A, S253A, and S316A mutants were ubiquitinated significantly lower than wild-type FKHR. Remarkably, the double mutants T24A/S253A, T24A/S316A, and S253A/S316A and the triple mutant (3A) were much less ubiquitinated than the one-point mutations (Fig. 3C). These results indicate that PKB-dependent phosphorylation plays a significant role for FKHR ubiquitination.
Fig. 3.
Phosphorylation via PI3K-PKB pathway is important for FKHR ubiquitination. (A) HepG2 cells were serum-starved for 12 h and preincubated with PI3K inhibitor, LY294002, or wortmannin for 30 min and stimulated with insulin for 6 h. Whole cell extracts were analyzed by immunoblotting using anti-FKHR antibody. (B) FLAG-FKHR and HA-ubiquitin were cotransfected with a dominant negative mutant of PI3K (Xp-Δp85) expression plasmids in HepG2 cells. After treatment with MG132, whole cell extracts were subjected to anti-FLAG immunoprecipitation and followed by immunoblotting. The expression of Xpress-tagged Δp85 in the cell extract was shown by immunoblotting with anti-Xpress antibody. (C) HepG2 cells were transfected with the indicated FKHR mutant and HA-ubiquitin expression plasmids and treated with MG132. Whole cell extracts were subjected to anti-FLAG immunoprecipitation, followed by anti-HA(Upper) or anti-FLAG (Lower) immunoblotting. (D) Cell extracts from HEK293T cells transfected with the FLAG-FKHR WT or 3A plasmid were subjected to anti-FLAG immunoprecipitation. The immune complexes were incubated at 30°C for 1 h in the presence or absence of RRL. The reaction products were analyzed by immunoblotting with anti-FKHR antibody.
We further assessed the necessity of phosphorylation for FKHR ubiquitination by an in vitro ubiquitination assay using wild-type FLAG-FKHR or the 3A mutant that is immunoprecipitated from transfected HEK293T cell extracts. As illustrated in Fig. 3D, wild-type FKHR, but not the 3A mutant, was ubiquitinated in the presence of RRL, indicating that the phosphorylated state is required for FKHR ubiquitination.
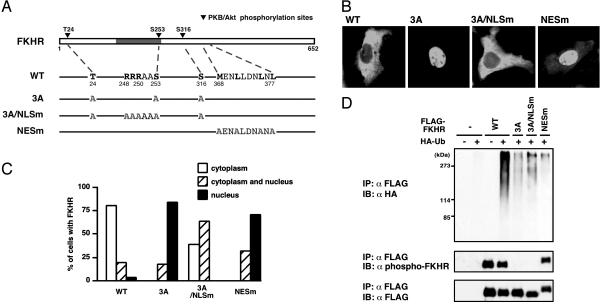
Effect of Subcellular Localization on FKHR Ubiquitination. It has been shown that FOXO proteins are phosphorylated by insulin in the nucleus and exported to the cytoplasm (31, 32). Because we provide evidence that ubiquitination of FKHR requires its phosphorylation in vivo and in vitro, these observations contain several possibilities. First, phosphorylated FKHR is ubiquitinated in the nucleus and transported to the cytoplasm. Second, accumulation of phosphorylated FKHR in the cytoplasm promotes FKHR ubiquitination. Third, phosphorylated FKHR is ubiquitinated regardless of the subcellular localization. To distinguish these possibilities, we investigated the effect of the subcellular localization on FKHR ubiquitination by using the FKHR mutants (Fig. 4A): one was 3A/NLSm, in which both three PKB phosphorylation sites and arginine residues important for nuclear localization (31–33) were replaced by alanines; the other was NESm, in which the critical leucine and methionine residues in NES (8, 32) were converted to alanines. In the presence of serum, wild-type FKHR was localized in the cytoplasm, whereas 3A mutant was in the nucleus (Fig. 4 B and C). Remarkably, 3A/NLSm mutant was located in the cytoplasm, despite PKB-unphosphorylated form, in a major population of transfected cells. In contrast, NESm mutant was predominantly localized in the nucleus, although this mutant was phosphorylated (Fig. 4 B and C, and D Middle). By using these mutants, we examined ubiquitination of FKHR in vivo. Both 3A/NLSm and NESm mutants, similar to 3A mutant, were much less ubiquitinated than wild-type (Fig. 4D). These results suggest that, in addition to the “phosphorylated” condition, the cytoplasmic retention is also necessary for efficient ubiquitination of FKHR.
Fig. 4.
Both phosphorylation and cytoplasmic retention are necessary for efficient FKHR ubiquitination. (A) A schematic representation of FKHR point mutants. The gray box indicates the forkhead DNA binding domain. (B) Localization of FKHR WT or mutants was monitored by transfection of these FKHRs into HepG2 cells in the presence of FBS and by immunolocalization with anti-FLAG antibody. (C) For quantitation, 100 cells per coverslips were counted, and the results are shown as the percentage of cells. (D) HepG2 cells were transfected with the indicated FKHR mutant and HA-ubiquitin expression plasmids and treated with MG132 in the presence of FBS. Whole cell extracts were subjected to anti-FLAG immunoprecipitation and followed by anti-HA (Top), anti-phospho-FKHR (Middle), or anti-FLAG (Bottom) immunoblotting.
Discussion
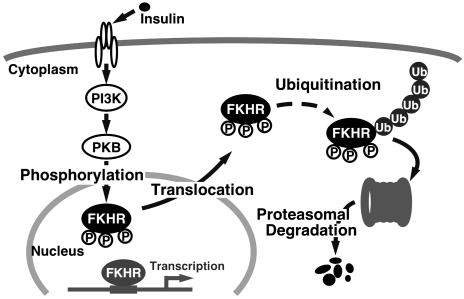
In this study, we have shown the degradation of FKHR phosphorylated by the PI3K-PKB pathway in response to insulin through the ubiquitin-proteasome system. Based on this finding, we speculate a mechanism for the regulation of FKHR by insulin (Fig. 5). When insulin binds to its specific receptor and activates the downstream signaling cascade, FKHR is phosphorylated by PKB and consequently excluded from the nucleus. Phosphorylated FKHR is then ubiquitinated in the cytoplasm, followed by degradation through the 26S proteasome. Recently, Plas and Thompson (34) have found that the expression of constitutively active PKB in hematopoietic cell line promotes proteasomemediated decline of several PKB substrates, including FOXO1 (FKHR) and FOXO3 (FKHRL1). Our striking finding in the present study is that insulin triggers FKHR ubiquitination and proteasome-mediated degradation that require both phosphorylation and cytoplasmic retention.
Fig. 5.
A model for FKHR regulation through insulin-induced and phosphorylation-dependent degradation.
The ubiquitin-proteasome system is involved in the regulation of a variety of basic cellular processes. Ubiquitination is catalyzed through a multienzyme cascade, including the ubiquitinactivating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin ligase (E3) (30). The E3 enzymes accomplish the last and most essential step in the conjugation reaction with substrate specificity, and in several cases the target proteins are first marked by phosphorylation for ubiquitination. For example, IκB and β-catenin are phosphorylated by IκB-kinase complex and glycogen synthate kinase-3β, respectively, and then ubiquitinated by an Skp1/cullin-1/F-box protein: β-TrCP (SCFβ-TrCP), which selectively binds to phosphorylated serine residues of the substrates (35). In the case of FKHR, a strong correlation between phosphorylation and ubiquitination prompted us to consider the biochemical significance of phosphorylation as a prerequisite for ubiquitination. The first possibility is that an E3 enzyme may specifically require the phosphorylated residue(s) of FKHR to recognize and bind to it as a target. Second, the phosphorylation may induce conformational changes of FKHR, leading to exposure of the E3-binding domain, and thus permitting an E3 enzyme access to FKHR. Third, 14–3-3 proteins, which can associate with phosphorylated FOXO proteins (5, 32, 33), may affect the FKHR ubiquitination by mediating the interaction with an E3 enzyme.
Recent studies have demonstrated that subcellular localization also contributes to protein ubiquitination. Smad2 protein is phosphorylated and activated by transforming growth factor-β receptors and translocated to the nucleus, where it controls transcription. Activated Smad2 is destroyed through the ubiquitin–proteasome pathway, in which its accumulation in the nucleus leads to ubiquitination mediated by Smurf2, whereas the receptor-phosphorylated region per se does not serve as a signal for ubiquitin conjugation (36–38). The p53 tumor suppressor protein levels are kept low during growth of normal cells through ubiquitination-mediated degradation, which is controlled by MDM2. p53 is ubiquitinated in the nucleus, exported to the cytoplasm, and degraded through proteasome (39). Our results indicate that insulin-induced export from the nucleus into the cytoplasm as well as phosphorylation is important for efficient FKHR ubiquitination, providing a novel mechanism of ubiquitination of proteins shuttling between the nucleus and the cytoplasm.
Given the physiological importance of insulin-dependent ubiquitination, the selective and irreversible degradation of phosphorylated FKHR subsequent to nuclear export may prevent the reentry of FKHR into the nucleus and principally contribute to sustaining the inhibitory effect of insulin on gene expression. Therefore, further studies to identify the E3 enzyme for FKHR would be expected to provide new insights into the mechanism by which insulin regulates FKHR function and gene expression.
Acknowledgments
We especially thank Dr. Wataru Ogawa, Kobe University, for the gift of the plasmid (pGEX-Δp85). We also thank the Fukamizu laboratory members for their helpful discussion and encouragement. This work was supported by The 21st Century COE Program, Grant-in-Aid for Scientific Research (A), and Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Technology of Japan, “Research for the Future” Program (The Japan Society for the Promotion of Science JSPS-RFTF 97L00804), the Research Grant for Cardiovascular Diseases (11C-1), Comprehensive Research on Aging and Health from the Ministry of Health, Labour, and Welfare, TAKEDA Science Foundation, and The Cell Science Research Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PKB, protein kinase B; PI3K, phosphatidylinositol 3-kinase; NLS, nuclear localization signal; NES, nuclear export signal; HA, hemagglutinin; RRL, rabbit reticulocyte lysate.
References
- 1.Kandel, E. S. & Hay, N. (1999) Exp. Cell Res. 253, 210–229. [DOI] [PubMed] [Google Scholar]
- 2.Kido, Y., Nakae, J. & Accili, D. (2001) J. Clin. Endocrinol. Metab. 86, 972–979. [DOI] [PubMed] [Google Scholar]
- 3.Burgering, B. M. & Kops, G. J. (2002) Trends Biochem. Sci. 27, 352–360. [DOI] [PubMed] [Google Scholar]
- 4.Birkenkamp, K. U. & Coffer, P. J. (2003) Biochem. Soc. Trans. 31, 292–297. [DOI] [PubMed] [Google Scholar]
- 5.Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., Anderson, M. J., Arden, K. C., Blenis, J. & Greenberg, M. E. (1999) Cell 96, 857–868. [DOI] [PubMed] [Google Scholar]
- 6.Kops, G. J., de Ruiter, N. D., De Vries-Smits, A. M., Powell, D. R., Bos, J. L. & Burgering, B. M. (1999) Nature 398, 630–634. [DOI] [PubMed] [Google Scholar]
- 7.Rena, G., Guo, S., Cichy, S. C., Unterman, T. G. & Cohen, P. (1999) J. Biol. Chem. 274, 17179–17183. [DOI] [PubMed] [Google Scholar]
- 8.Biggs, W. H., III, Meisenhelder, J., Hunter, T., Cavenee, W. K. & Arden, K. C. (1999) Proc. Natl. Acad. Sci. USA 96, 7421–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takaishi, H., Konishi, H., Matsuzaki, H., Ono, Y., Shirai, Y., Saito, N., Kitamura, T., Ogawa, W., Kasuga, M., Kikkawa, U. & Nishizuka, Y. (1999) Proc. Natl. Acad. Sci. USA 96, 11836–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayala, J. E., Streeper, R. S., Desgrosellier, J. S., Durham, S. K., Suwanichkul, A., Svitek, C. A., Goldman, J. K., Barr, F. G., Powell, D. R. & O'Brien, R. M. (1999) Diabetes 48, 1885–1889. [DOI] [PubMed] [Google Scholar]
- 11.Schmoll, D., Walker, K. S., Alessi, D. R., Grempler, R., Burchell, A., Guo, S., Walther, R. & Unterman, T. G. (2000) J. Biol. Chem. 275, 36324–36333. [DOI] [PubMed] [Google Scholar]
- 12.Hall, R. K., Yamasaki, T., Kucera, T., Waltner-Law, M., O'Brien, R. & Granner, D. K. (2000) J. Biol. Chem. 275, 30169–30175. [DOI] [PubMed] [Google Scholar]
- 13.Daitoku, H., Yamagata, K., Matsuzaki, H., Hatta, M. & Fukamizu, A. (2003) Diabetes 52, 642–649. [DOI] [PubMed] [Google Scholar]
- 14.Nakae, J., Biggs, W. H., III, Kitamura, T., Cavenee, W. K., Wright, C. V., Arden, K. C. & Accili, D. (2002) Nat. Genet. 32, 245–253. [DOI] [PubMed] [Google Scholar]
- 15.Taub, J., Lau, J. F., Ma, C., Hahn, J. H., Hoque, R., Rothblatt, J. & Chalfie, M. (1999) Nature 399, 162–166. [DOI] [PubMed] [Google Scholar]
- 16.Honda, Y. & Honda, S. (1999) FASEB J. 13, 1385–1393. [PubMed] [Google Scholar]
- 17.Ishii, N., Goto, S. & Hartman, P. S. (2002) Free Radical Biol. Med. 33, 1021–1025. [DOI] [PubMed] [Google Scholar]
- 18.Nemoto, S. & Finkel, T. (2002) Science 295, 2450–2452. [DOI] [PubMed] [Google Scholar]
- 19.Kops, G. J., Dansen, T. B., Polderman, P. E., Saarloos, I., Wirtz, K. W., Coffer, P. J., Huang, T. T., Bos, J. L., Medema, R. H. & Burgering, B. M. (2002) Nature 419, 316–321. [DOI] [PubMed] [Google Scholar]
- 20.Zhao, H. H., Herrera, R. E., Coronado-Heinsohn, E., Yang, M. C., Ludes-Meyers, J. H., Seybold-Tilson, K. J., Nawaz, Z., Yee, D., Barr, F. G., Diab, S. G., et al. (2001) J. Biol. Chem. 276, 27907–27912. [DOI] [PubMed] [Google Scholar]
- 21.Schuur, E. R., Loktev, A. V., Sharma, M., Sun, Z., Roth, R. A. & Weigel, R. J. (2001) J. Biol. Chem. 276, 33554–33560. [DOI] [PubMed] [Google Scholar]
- 22.Hirota, K., Daitoku, H., Matsuzaki, H., Araya, N., Yamagata, K., Asada, S., Sugaya, T. & Fukamizu, A. (2003) J. Biol. Chem. 278, 13056–13060. [DOI] [PubMed] [Google Scholar]
- 23.Hatta, M., Daitoku, H., Matsuzaki, H., Deyama, Y., Yoshimura, Y., Suzuki, K., Matsumoto, A. & Fukamizu, A. (2002) Int. J. Mol. Med. 9, 147–152. [PubMed] [Google Scholar]
- 24.Ebisawa, T., Fukuchi, M., Murakami, G., Chiba, T., Tanaka, K., Imamura, T. & Miyazono, K. (2001) J. Biol. Chem. 276, 12477–12480. [DOI] [PubMed] [Google Scholar]
- 25.Kotani, K., Yonezawa, K., Hara, K., Ueda, H., Kitamura, Y., Sakaue, H., Ando, A., Chavanieu, A., Calas, B., Grigorescu, F., et al. (1994) EMBO J. 13, 2313–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Hoog, C. L., Koehler, J. A., Goldstein, M. D., Taylor, P., Figeys, D. & Moran, M. F. (2001) Mol. Cell. Biol. 21, 2107–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagano, M., Tam, S. W., Theodoras, A. M., Beer-Romero, P., Del Sal, G., Chau, V., Yew, P. R., Draetta, G. F. & Rolfe, M. (1995) Science 269, 682–685. [DOI] [PubMed] [Google Scholar]
- 28.Conaway, R. C., Brower, C. S. & Conaway, J. W. (2002) Science 296, 1254–1258. [DOI] [PubMed] [Google Scholar]
- 29.Desterro, J. M., Rodriguez, M. S. & Hay, R. T. (2000) Cell. Mol. Life Sci. 57, 1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weissman, A. M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 169–178. [DOI] [PubMed] [Google Scholar]
- 31.Brownawell, A. M., Kops, G. J., Macara, I. G. & Burgering, B. M. (2001) Mol. Cell. Biol. 21, 3534–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunet, A., Kanai, F., Stehn, J., Xu, J., Sarbassova, D., Frangioni, J. V., Dalal, S. N., DeCaprio, J. A., Greenberg, M. E. & Yaffe, M. B. (2002) J. Cell. Biol. 156, 817–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rena, G., Prescott, A. R., Guo, S., Cohen, P. & Unterman, T. G. (2001) Biochem. J. 354, 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plas, D. R. & Thompson, C. B. (2003) J. Biol. Chem. 278, 12361–12366. [DOI] [PubMed] [Google Scholar]
- 35.Maniatis, T. (1999) Genes Dev. 13, 505–510. [DOI] [PubMed] [Google Scholar]
- 36.Lo, R. S. & Massague, J. (1999) Nat. Cell Biol. 1, 472–478. [DOI] [PubMed] [Google Scholar]
- 37.Lin, X., Liang, M. & Feng, X. H. (2000) J. Biol. Chem. 275, 36818–36822. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, Y., Chang, C., Gehling, D. J., Hemmati-Brivanlou, A. & Derynck, R. (2001) Proc. Natl. Acad. Sci. USA 98, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michael, D. & Oren, M. (2003) Semin. Cancer Biol. 13, 49–58. [DOI] [PubMed] [Google Scholar]