Abstract
Intron-containing genes are generally expressed more effectively in human cells than are intronless versions of the same gene. We have asked whether this effect is due directly to splicing or instead reflects the action of components of the exon junction complex (EJC) that is assembled at splice junctions after splicing is completed. Here, we show that intron removal does not enhance gene expression if EJC formation is blocked. Conversely, RNA tethering of the EJC components SRm160 or RNPS1 boosts the expression of intronless mRNAs but not of spliced mRNAs. Splicing and RNPS1 tethering are shown to enhance the same steps in mRNA biogenesis and function, including mRNA 3′ end processing and translation. Together, these data argue that the EJC is primarily responsible for the positive effect of splicing on gene expression.
Almost all metazoan genes contain introns that must be precisely removed from the initial pre-mRNA before nuclear export and translation. However, intron removal not only renders an mRNA suitable for cytoplasmic utilization but also enhances the level of expression of the encoded protein when compared with an intronless version of the same gene (1–4). This stimulatory effect, which can vary from a modest ≈2-fold to >30 fold, depending on the gene and cell being examined, results from effects of intron removal on several steps in the mRNA life cycle. Thus, splicing has been reported to enhance mRNA transcription and 3′ end processing, to increase nuclear and cytoplasmic mRNA levels, to promote nuclear mRNA export, and to enhance mRNA translational utilization (1–8).
Removal of an intron not only modifies the primary sequence of an mRNA but also tags the mRNA with a protein complex of ≈335 kDa, termed an exon junction complex (EJC) (9–11). The EJC, which is deposited ≈20–24 nt 5′ of the splice junction, provides a position-specific memory of the splicing event. Indeed, several proteins that form part of the EJC remain associated, at least transiently, with not only nuclear but also cytoplasmic spliced mRNAs (9, 12, 13).
Analysis of the EJC has identified several protein components, including SRm160, RNPS1, Aly/REF, Y14, and Magoh (9–11). Both SRm160 and RNPS1 can function as coactivators of splicing, and SRm160 has also been proposed to enhance mRNA 3′ end processing (14, 15). Both RNPS1 and Y14 have been implicated in the process of nonsense-mediated decay, which, in higher eukaryotes, depends on the EJC (12, 16). Magoh may facilitate the subcytoplasmic localization of mRNAs (17), whereas Aly has been implicated in nuclear mRNA export (18, 19). Aly is thought to be recruited to the EJC by the splicing factor UAP56 and is then believed to interact with the Tap nuclear mRNA export factor (reviewed in ref. 20). However, unlike both Tap and UAP56, Aly is dispensable for bulk mRNA export in metazoan cells (21–23). This result may be explained by recent data arguing that specific serine/arginine-rich splicing factors can also function as adapter proteins for Tap-dependent mRNA export (24).
In this article, we have asked whether the stimulatory effect of mRNA splicing on gene expression is mediated by the process of splicing itself or is instead, or in addition, due to the action of components of the EJC. Our data demonstrate that splicing fails to enhance gene expression if formation of the EJC is prevented. In contrast, mRNA tethering of the EJC components SRm160 and RNPS1 enhances the expression of intronless mRNAs but not of analogous intron-containing transcripts. Tethering of RNPS1 facilitated the 3′ end processing of mRNAs and enhanced the translational utilization of bound mRNAs but did not appear to affect nuclear mRNA export. Together, these data suggest that the stimulatory effect of splicing on gene expression may be largely or entirely due to the action of components of the EJC.
Methods
Construction of Molecular Clones. All expression plasmids used in this work are based on pBC12/CMV (CMV, cytomegalovirus). The plasmids have been described as follows: pCMV/I-β-gal (β-gal, β-galactosidase), pCMV/CAT (CAT, chloramphenicol acetyltransferase), pCMV/I-CAT, pCMV/WT/GLB, pCMV/Δ1+2/GLB, pCMV/I/Δ1+2/GLB, pCMV/Δ1+2/GLB/CTE, pCMV/Δ1+2/GLB/RRE, and pDM128/PL (2, 25). pCMV/N was generated by annealing sense primer 1 (5′-CATGGGCAATGCTAAAACTCGCCGTCATGAGCGGCGCAGAAAGCTAGCCATAGAGCGCGACACCATCGAATTCGATCC-3′) and antisense primer 2 (5′-TCGAGGATCGAATTCGATGGTGTCGCGCTCTATGGCTAGCTTTCTGCGCCGCTCATGACGGCGAGTTTTAGCATTGCC-3′) and then ligating the resultant double-stranded product into pBC12/CMV cut with NcoI and XhoI. This oligonucleotide encodes the bacteriophage P22 N-peptide sequence NH2-NAKTRRHERRRKLAIERDTI-COOH (26) flanked at the amino terminus by the residues Met-Gly. The introduced EcoRI site and the existing XhoI site were then used to insert the different EJC proteins as well as UAP56, Tap, and Rev. RNPS1, Y14, and UAP56 were obtained as EcoRI/XhoI fragments by PCR from a human cDNA library. SRm160, Aly, Tap, and Rev were cloned from described expression plasmids (2, 15, 25).
pCMV/CAT/B, pCMV/I-CAT/B, pDM128/B, and pCMV/Δ1 + 2/GLB/B were constructed by ligating in annealed oligonucleotides. Primer 3 (sense), 5′-TCGACTAGGCGCTGACAAAGCGCCATGACTAGGCGCTGACAAAGCGCCCTAC-3′, and primer 4 (antisense), 5′-TCGAGTAGGGCGCTTTGTCAGCGCCTAGTCATGGCGCTTTGTCAGCGCCTAG-3′, containing two phage P22 box-B sequences, were annealed and cloned into the unique XhoI site present in the 3′ UTR of all of the above indicator plasmids. The annealed oligonucleotides were inserted three times, resulting in six box-B sequences. The 5-nt box-B RNA terminal loops are underlined, and each is flanked by a 6-bp inverted repeat predicted to form an RNA stem. pCMV/SX/Δ1+2/GLB (SX, short exon) was generated by partial deletion of the 5′ UTR exon present in pCMV/I/Δ1+2/GLB by using PCR. pCMV/SX/WT/GLB was constructed by replacement of the β-globin cDNA present in pCMV/SX/Δ1+2/GLB with the genomic β-globin gene bearing two introns.
Reporter Assays. Human 293T cells were maintained as described (25) and were transfected by using FuGENE 6 (Roche Molecular Biochemicals) or calcium phosphate. Induced CAT enzyme activities were determined ≈48 h after transfection and were adjusted for minor differences in transfection efficiency or sample recovery, as revealed by a cotransfected β-gal internal control plasmid. Western blot assays and quantitation of reactive protein levels were performed as described (2).
RNA Isolation and RNase Protection Assay (RPA). Nuclear and cytoplasmic RNA isolation and RPAs were performed as described (2). RNA probes were generated by in vitro transcription of PCR products containing a T7 promoter sequence, by using a Riboprobe kit (Promega). The RNA probe that traverses the preproinsulin genomic polyadenylation site has been described (2). The RNA probe that traverses the insulin 5′ UTR intron 3′ splice site is 365 nt in length, whereas probe fragments rescued by unspliced and spliced mRNAs are predicted to be 335 and 211 nt, respectively. Protected probe fragments were separated by gel electrophoresis, visualized by autoradiography, and quantified by using a PhosphorImager.
Results
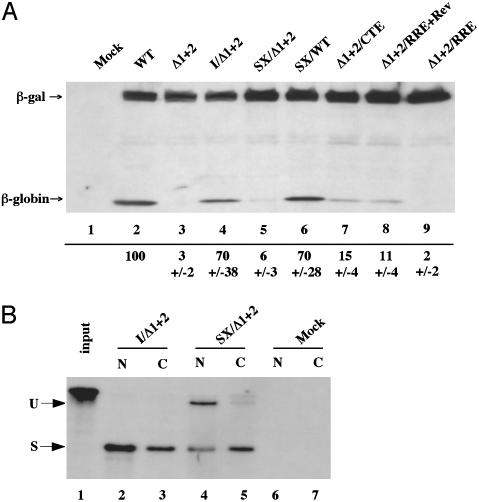
EJC Recruitment Enhances mRNA Utilization. The human β-globin gene is unusually dependent on splicing for effective protein expression (1, 2). This result is reproduced in Fig. 1A, which shows that a described (2) expression plasmid containing the genomic β-globin gene, which contains two introns within the ORF (pCMV/WT/GLB, lane 2), expresses ≥30-fold more hemagglutinin epitope-tagged β-globin in transfected 293T cells than does an expression plasmid that is identical except that the two introns have been removed (pCMV/Δ1+2/GLB, lane 3).
Fig. 1.
The positive effect of splicing on β-globin gene expression depends on EJC recruitment. (A) Western analysis of the level of hemagglutinin-β-globin expression observed in 293T cells transfected with the indicated expression plasmids. Cells were transfected with 400 ng of each β-globin expression plasmid and 100 ng of pCMV/I-β-gal, which served as an internal control. This panel shows a representative experiment with quantitative data derived from three independent experiments, complete with standard deviation, given below each lane. (B) RPA of nuclear (N) and cytoplasmic (C) RNA fractions derived from 293T cells transfected with pCMV/I/Δ1+2/GLB or pCMV/SX/Δ1+2/GLB, or mock transfected. The probe used traverses the 3′ splice site of the 5′ UTR intron and therefore measures the level of expression of both unspliced (U) and spliced (S) mRNA.
The 5′ UTR used in pCMV/WT/GLB and pCMV/Δ1+2/GLB is derived from the rat preproinsulin II gene, and this 5′ UTR, in its genomic form, contains an intron (2). Substitution of the intron-containing form of the 5′ UTR, in place of the cDNA version, into pCMV/Δ1+2/GLB rescued β-globin protein expression (Fig. 1, lane 4). The 5′ noncoding exon present in the resultant pCMV/I/Δ1+2/GLB plasmid is predicted to be 122 nt in length, and this is clearly sufficient to accommodate the EJC, which is deposited 20–24 nt 5′ to the splice junction. LeHir et al. (10) have demonstrated, however, that a 5′ exon of 17 nt is inadequate to support EJC formation. Truncation of the 5′ exon present in pCMV/I/Δ1+2/GLB to 17 nt, to give pCMV/SX/Δ1+2/GLB, resulted in a marked drop in the level of β-globin protein expression (Fig. 1 A, lane 5). To test whether truncation of the 5′ UTR reduced β-globin expression for reasons unrelated to EJC formation, we also substituted this 5′ SX into the genomic β-globin expression plasmid pCMV/WT/GLB, to give pCMV/SX/WT/GLB. As shown in Fig. 1, lane 6, this plasmid, which is predicted to encode a mature mRNA identical to pCMV/SX/Δ1+2/GLB, gave rise to high levels of β-globin.
It is important to confirm that the intron introduced into the 5′ UTR of both pCMV/I/Δ1+2/GLB and pCMV/SX/Δ1+2/GLB is actually spliced. We therefore prepared nuclear and cytoplasmic RNA fractions and analyzed the level of unspliced and spliced transcripts by RPA. As shown in Fig. 1B, lanes 2 and 3, we detected only spliced transcripts in both the nuclear and cytoplasmic fraction of cells transfected with the pCMV/I/Δ1+2/GLB. In contrast, cells transfected with the plasmid bearing the truncated, 17-nt 5′ exon gave rise to readily detectable levels of both spliced and unspliced mRNA in the nuclear fraction (Fig. 1B, lane 4) yet also gave rise to almost exclusively spliced cytoplasmic mRNA (lane 5). It therefore appears that truncation of the 5′ exon from 122 to 17 nt reduces the efficiency of splicing but that mRNA transcripts were spliced before nuclear export. Nevertheless, very little β-globin protein was expressed (Fig. 1 A, lane 5).
Tethering of EJC Components Rescues Intronless mRNA Expression. The data presented in Fig. 1 suggest that EJC recruitment is necessary for the enhancement of gene expression induced by splicing. We next asked whether tethering components of the EJC to an intronless mRNA would suffice to rescue mRNA utilization. For this purpose, we constructed an indicator construct, termed pCMV/CAT/B, consisting of the CMV immediate early promoter driving a cat gene linked in turn to six copies of an RNA-binding site, termed a “box B,” derived from the lambdoid phage P22 (26) (Fig. 2D). This RNA stem–loop serves as the binding site for a 20-aa RNA-binding domain termed the N-peptide. DeGregorio et al. (27) have shown, using similar box-B and N-peptide sequences derived from phage λ, that this interaction can support the effective recruitment of fusion proteins to box-B RNA target sites in human cells.
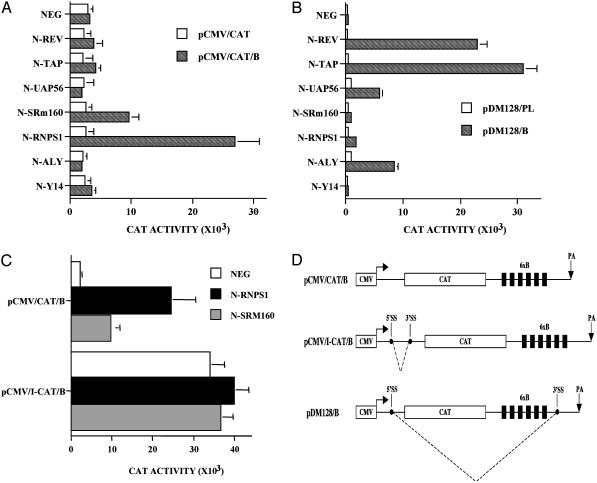
Fig. 2.
Tethered EJC proteins can activate expression of an intronless gene. (A) 293T cells were transfected with 100 ng of the pCMV/CAT/B or pCMV/CAT indicator plasmid and 300 ng of a plasmid encoding an N-peptide fusion protein, or pBC12/CMV as a negative (NEG) control. Cells were also cotransfected with 25 ng of pCMV/I-β-gal as an internal control. Induced CAT activities were measured at 48 h posttransfection and adjusted for minor differences in the β-gal internal control. Data shown represent the average of three experiments with standard deviation indicated. (B) This assay was performed as shown for A except that cells were transfected with 20 ng of the pDM128/B or pDM128/PL indicator plasmid. (C) This assay was performed as shown in A by using either the intronless pCMV/CAT/B or the intron-containing pCMV/I-CAT/B indicator plasmid. (D) Schematic representation of the indicator plasmids used in A–C.
Although the cat gene is of prokaryotic origin, we have observed that CAT protein expression is increased 10- to 15-fold in 293T cells when an intron is present in the 5′ UTR (2) (Fig. 2C). To test whether tethering of EJC components might exert a similar enhancing effect, we constructed plasmids designed to express a range of fusion proteins consisting of the P22 N-peptide attached to the amino terminus of the EJC components SRm160, RNPS1, Aly, Y14, and Magoh. We also analyzed fusion proteins consisting of the N-peptide fused to UAP56, which is involved in Aly recruitment to the EJC (28) and Tap, a human mRNA export factor that can interact with the EJC (10). A final factor included in this analysis is the HIV type 1 (HIV-1) Rev protein, which functions as a Crm-1-dependent nuclear mRNA export factor (20). Western analysis of 293T cells transfected with each expression plasmid (data not shown) revealed a readily detectable level of expression of all fusion proteins except N-Magoh, which was therefore excluded from further analysis.
Cotransfection of each N-peptide fusion protein expression plasmid with pCMV/CAT/B, or with a similar control vector lacking box-B sequences (pCMV/CAT), revealed an ≈10-fold activation of CAT expression by N-RNPS1 and an ≈4-fold activation by N-SRm160, both of which were dependent on the presence of B boxes (Fig. 2 A). No significant activation (≤2-fold) was noted on cotransfection with N-Rev, N-Tap, N-Y14, N-UAP56, or N-Aly.
A striking aspect of the data shown in Fig. 2 A is that factors believed to function in nuclear mRNA export, i.e., Tap, Aly, UAP56, and Rev, had little or no enhancing effect on the level of CAT expression when tethered to mRNAs transcribed from pCMV/CAT/B. We therefore wished to confirm that these fusion proteins were indeed capable of mediating the nuclear export of tethered mRNAs. For this purpose, we constructed a derivative of the pDM128/PL indicator plasmid (25), termed pDM128/B, containing six 3′ UTR box-B sequences (Fig. 2D). The pDM128/PL indicator construct and its derivatives represent a well established tool for determining the nuclear RNA export potential of proteins recruited either by tethering or by their cognate RNA target (25, 29, 30). As shown in Fig. 2D, pDM128/B contains a cat gene sequestered between 5′ and 3′ splice sites derived originally from HIV-1. The unspliced mRNA transcribed from pDM128/B, which encodes CAT, is normally unable to be exported from the nucleus because of nuclear retention induced by the splice sites present in cis (20, 31). However, if a nuclear mRNA export factor is recruited to this unspliced mRNA, then nuclear export, and CAT enzyme expression, can be detected. Several groups have shown that CAT activity is an accurate measure of the cytoplasmic level of this unspliced mRNA (25, 29, 30).
Cotransfection of each N-peptide fusion protein with either pDM128/B or the pDM128/PL control plasmid confirmed the reported ability of tethered HIV-1 Rev or human Tap to induce expression of the unspliced CAT mRNA (25, 30) (Fig. 2B). In addition, we also observed that both N-UAP56 and N-Aly were able to induce a significant level of CAT expression when coexpressed with pDM128/B, consistent with their proposed role in nuclear mRNA export. In contrast, neither N-RNPS1 nor N-SRm160 activated CAT expression from pDM128/B (Fig. 2B). Therefore, there is a complete lack of concordance between the ability of the tested proteins to activate CAT expression from pDM128/B versus from pCMV/CAT/B, with known nuclear mRNA export factors being able to activate only the former. As a further test of whether bound nuclear mRNA export factors would be able to rescue the expression of intronless mRNAs, we inserted either the constitutive transport element (CTE), a physiological RNA target for Tap, or the Rev response element (RRE), the normal RNA target for HIV-1 Rev, into the 3′ UTR of the intronless pCMV/Δ1+2/GLB construct. Although both of these RNA export elements gave rise to an ≈4-fold increase in the level of β-globin protein expression (Fig. 1 A, lanes 7 and 8), this effect was clearly far less than the 23- to 33-fold increase seen on inclusion of an intron in cis. Therefore, we conclude that inefficient nuclear mRNA export is not the major limiting step causing poor expression of intronless mRNAs.
If the tethered RNPS1 and SRm160 proteins are indeed rescuing expression of the cat mRNA encoded by pCMV/CAT/B by mimicking formation of the EJC during splicing, then one would predict that insertion of an intron would render these fusion proteins irrelevant. Conversely, if RNPS1 and SRm160 are acting by mechanisms unrelated to EJC function, then one might predict functional synergy. To test whether RNPS1 and SRm160 recruitment by tethering and by EJC formation are indeed functionally redundant, we inserted an intron into the 5′ UTR of pCMV/CAT/B. The 5′ UTR present in pCMV/CAT/B is, in fact, the same preproinsulin 5′ UTR used in the β-globin expression plasmids analyzed in Fig. 1, and we therefore simply substituted the genomic, intron-containing form of this 5′ UTR in place of the cDNA version, to generate pCMV/I-CAT/B (Fig. 2D). As shown in Fig. 2C, and previously reported (2), this intron enhances CAT expression by 10- to 15-fold. Importantly, however, the presence of this intron also renders pCMV/I-CAT/B refractory to the enhancing effect of both N-RNPS1 and N-SRm160. This result is consistent with the hypothesis that tethering of N-RNPS1 or N-SRm160 is indeed mimicking the biological activity of the EJC.
We note that tethering of RNPS1 also did not inhibit CAT expression, as might be predicted based on earlier work showing that tethering of RNPS1 to an mRNA can induce nonsense-mediated decay (12). Although we do not know why no inhibitory effect was observed in these experiments, we note that the box-B RNA-binding sites were inserted starting 6 nt 3′ to the CAT translation termination codon in pCMV/I-CAT/B. In the case of nonsense-mediated decay caused by a 3′ UTR intron, it is known that close proximity to the translation termination codon prevents mRNA degradation (32)
Tethering of RNPS1 to an Intronless β-Globin mRNA Enhances 3′ End Processing and Translational Utilization. The data presented in Fig. 2 demonstrate that tethering of RNPS1 or SRm160 to an intronless CAT mRNA, but not to an intron-containing CAT mRNA, significantly enhances CAT enzyme expression in transfected human cells. We next wished to extend this observation to a human gene, i.e., the β-globin gene, and to analyze this effect at the mRNA level.
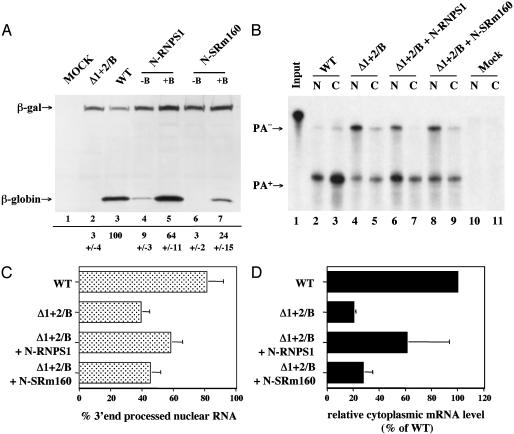
The pCMV/Δ1+2/B/GLB plasmid is identical to the intronless pCMV/Δ1+2/GLB plasmid analyzed in Fig. 1 except that it contains six box-B targets inserted into the 3′ UTR. This insertion did not, in and of itself, affect the level of β-globin protein expression (Fig. 3A, lane 2). To test whether coexpression of either N-RNPS1 or N-SRm160 would enhance β-globin protein expression, we cotransfected pCMV/Δ1+2/B/GLB, or pCMV/Δ1+2/GLB as a negative control, with the relevant N-peptide fusion protein expression plasmids. As shown in Fig. 3A, coexpression of N-RNPS1 enhanced the level of β-globin expression from pCMV/Δ1+2/B/GLB by ≈21-fold (lane 5), whereas N-SRm160 enhanced β-globin expression by ≈8-fold (lane 7). Although N-SRm160 had no detectable effect on the expression of a similar intronless β-globin mRNA lacking box-B RNA targets (Fig. 3A, lane 6), we consistently observed a low, nonspecific enhancing effect of N-RNPS1 on the expression of this intronless mRNA (Fig. 3A, lane 4). This effect, which was also seen on coexpression of nonfused RNPS1 (data not shown), may suggest that the β-globin mRNA contains a low-affinity binding site(s) for RNPS1 that is not present in the intronless CAT mRNA analyzed in Fig. 2 A. Importantly, however, neither N-RNPS1 nor N-SRm160 affected the expression of the intron-containing β-gal mRNA used as an internal control (Fig. 3A).
Fig. 3.
Comparison of the effect of splicing and EJC protein tethering on intronless mRNA expression. (A) Western analysis of the effect of the N-RNPS1 or N-SRm160 fusion protein on the level of β-globin expression from an intronless expression plasmid (pCMV/Δ1 + 2/GLB) lacking (–B) or containing (+B) box-B binding sites. This panel shows a representative result with quantitative data drawn from three independent experiments summarized at the bottom. The pCMV/I-β-gal plasmid served as the internal control. (B) RPA of nuclear (N) and cytoplasmic (C) RNA fractions obtained from 293T cells transfected with the indicated indicator and effector plasmids. The probe used traverses the genomic preproinsulin 3′ polyadenylation site used and can therefore quantitate the level of β-globin transcripts that are (PA+) or are not (PA–) appropriately 3′ end processed. The effector (pCMV/N-RNPS1 and pCMV/N-SRm160) and control (pBC12/CMV) plasmids use a genomic bovine growth hormone polyadenylation site. (C) Data obtained from three independent RPAs, performed as shown in B, and the average steady-state level of appropriately 3′ end processed nuclear β-globin mRNA as a percentage of the total nuclear β-globin mRNA level. (D) Relative level of cytoplasmic β-globin mRNA, detected as described for B. The mRNA levels shown are normalized to the culture transfected with pCMV/WT/GLB, which is arbitrarily set at 100.
We have demonstrated that splicing enhances the expression of the β-globin mRNA encoded by pCMV/WT/GLB, when compared with the in principle identical β-globin mRNA encoded by pCMV/Δ1+2/GLB, in at least three ways. Specifically, splicing enhanced the efficiency of 3′ end processing, moderately increased the steady-state level of both nuclear and cytoplasmic β-globin mRNA, and also enhanced the cytoplasmic translational utilization of that mRNA (2). RPA data confirming this result are shown in Fig. 3B and quantified in Fig. 3 C and D. These data reveal that splicing of the genomic β-globin gene present in pCMV/WT/GLB increases correct 3′ end processing from ≈39% to ≈81% and increases cytoplasmic β-globin mRNA levels by ≈5-fold. This amount is, nevertheless, much less than the ≈33-fold increase in β-globin protein expression noted in Fig. 3A, thus also implying an effect on mRNA translation.
Analysis of the effect of the N-RNPS1 fusion protein on the expression of the intronless β-globin mRNA encoded by pCMV/Δ1+2/B/GLB revealed a similar phenotype. Thus, coexpression of N-RNPS1 significantly increased both 3′ end processing efficiency and cytoplasmic β-globin mRNA levels (Fig. 3 B–D). This ≈3-fold effect of N-RNPS1 tethering on the cytoplasmic β-globin mRNA level was, however, markedly lower than the ≈21-fold effect seen at the protein level (Fig. 3A). Analysis of N-SRm160 tethering, which had a relatively modest effect on β-globin protein expression (Fig. 3A), revealed a comparable modest positive effect at the mRNA level (Fig. 3 B–D).
Discussion
Intron-containing genes are generally expressed at a significantly higher level in human cells than the same genes lacking introns (1–4). Splicing may enhance gene expression at several, probably interconnected, steps including transcription, 3′ end processing, mRNA stability, nuclear RNA export, and finally translational utilization (1–8). The discovery of the EJC (9–11), and the realization that certain EJC proteins can accompany mature mRNAs to the cytoplasm (12, 13), has raised the possibility that splicing may exert this positive effect at least in part indirectly, i.e., by means of the EJC.
To test this hypothesis, we have analyzed the expression of spliced mRNAs that are either capable or incapable of recruiting an EJC (Fig. 1) and have also examined whether tethering of specific EJC proteins can rescue the expression of intronless mRNAs (Figs. 2 and 3). Our data demonstrate that splicing had no detectable positive effect on the expression of the highly intron-dependent human β-globin gene when the intron was positioned so close to the 5′ end of the mRNA that the EJC could not form, even though splicing of this mRNA still occurred (Fig. 1). In contrast, tethering of the EJC proteins SRm160 and, particularly, RNPS1 had a profound positive effect on the expression of intronless CAT and β-globin mRNAs (Figs. 2 A and 3A). Importantly, tethering did not enhance the expression of otherwise identical intron-containing mRNAs (Fig. 2C), thus strongly suggesting that these tethered EJC proteins are indeed mimicking the function of the authentic EJC. Together, these data strongly suggest that EJC recruitment accounts for most, and, at least in these experiments, possibly all, of the positive effect of splicing on gene expression in human cells.
Analysis of the steps affected by the tethered RNPS1 and SRm160 proteins demonstrates that RNPS1, in particular, acted by enhancing 3′ end processing, by increasing mRNA steadystate levels, and by increasing mRNA translational utilization, i.e., the same steps in the mRNA life cycle affected by splicing (2, 3). It is perhaps not surprising that the reported positive effect of splicing on mRNA translation appears to be due to EJC proteins. RNPS1, which has been implicated in nonsense-mediated decay (12) and must, therefore, be able to interact with the translational apparatus, is a reasonable candidate for this activity. The stimulatory effect of the EJC, and particularly of RNPS1 tethering, on mRNA 3′ end processing may be more surprising, given previous data reporting direct interactions between components of the basal splicing and 3′ mRNA processing machinery (7).
It is important to note that these tethering experiments do not directly address which component(s) of the EJC are actually responsible for the positive effects on mRNA expression that are observed. RNPS1 and SRm160 may well recruit other EJC components to the tethered mRNA, so that the observed phenotype would actually be indirect. We also note that RNPS1 and SRm160 interact specifically in the yeast two-hybrid assay (data not shown) so that tethering of either of these proteins may well result in recruitment of the other.
Although it has been suggested (5, 10, 33) that the EJC may play a critical role in the recruitment of nuclear mRNA export factors, including Tap, our data do not fully support this hypothesis. Tethering of proteins that function in nuclear mRNA export, i.e., HIV-1 Rev, UAP56, the EJC component Aly, and Tap itself, failed to enhance intronless mRNA expression (Fig. 2 A), even though all four of these proteins were able to induce nuclear export when tethered to an mRNA that was otherwise sequestered in the nucleus (Fig. 2B). In contrast, tethering of RNPS1 or SRm160 selectively enhanced the expression of intronless mRNAs (Figs. 2 A and 3A) yet failed to induce nuclear mRNA export (Fig. 2B). These data, when considered together with other recent reports (2, 3, 19, 21–23, 34), therefore suggest that the role of splicing and the EJC in mediating Tap recruitment and nuclear mRNA export is likely to be fairly modest and/or largely redundant with other steps in mRNA biogenesis. Nevertheless, even a fairly modest enhancement in mRNA export, due to more efficient recruitment of export factors to spliced mRNAs, could be very important if it affects essentially the entire cellular transcriptome.
If splicing and the resultant EJC play an important role in facilitating subsequent steps in the mRNA life cycle, then why are many mRNAs expressed quite effectively even in the absence of splicing? One possible explanation is that one or more EJC components may also be recruited to mRNAs independently of splicing, e.g., during transcription or because of interactions with low-affinity binding sites on the mRNA (20). Perhaps consistent with this hypothesis, overexpression of the N-RNPS1 fusion protein enhanced expression of not only an intronless β-globin mRNA containing box-B RNA targets but also of a similar mRNA lacking box-B sequences, although far more weakly in the latter case (Fig. 3A). It has in fact been suggested that proteins involved in EJC formation, such as UAP56, and components of the EJC, such as Aly, may be recruited to mRNAs independently of splicing (35, 36). Perhaps the ability of mRNAs to recruit specific EJC proteins independently of splicing correlates with their ability to be effectively expressed in an intronless form. In the most extreme version of this hypothesis, it seems possible that some mRNAs may have evolved high-affinity binding sites for one or more EJC components that allow these mRNAs to become entirely independent of splicing for efficient expression. It will therefore be interesting to examine whether any of the RNA sequences that have been shown to promote intronless mRNA expression (37) can specifically bind to components of the EJC.
Abbreviations: CAT, chloramphenicol acetyltransferase; CMV, cytomegalovirus; EJC, exon junction complex; β-gal, β-galactosidase; RPA, RNase protection assay; SX, short exon.
References
- 1.Buchman, A. R. & Berg, P. (1988) Mol. Cell. Biol. 8, 4395–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu, S. & Cullen, B. R. (2003) RNA 9, 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nott, A., Meislin, S. H. & Moore, M. J. (2003) RNA 9, 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryu, W.-S. & Mertz, J. E. (1989) J. Virol. 63, 4386–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo, M.-J. & Reed, R. (1999) Proc. Natl. Acad. Sci. USA 96, 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niwa, M., Rose, S. D. & Berget, S. M. (1990) Genes Dev. 4, 1552–1559. [DOI] [PubMed] [Google Scholar]
- 7.Vagner, S., Vagner, C. & Mattaj, I. W. (2000) Genes Dev. 14, 403–413. [PMC free article] [PubMed] [Google Scholar]
- 8.Furger, A., O'Sullivan, J. M., Binnie, A., Lee, B. A. & Proudfoot, N. J. (2002) Genes Dev. 16, 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kataoka, N., Yong, J., Kim, V. N., Velazquez, F., Perkinson, R. A., Wang, F. & Dreyfuss, G. (2000) Mol. Cell 6, 673–682. [DOI] [PubMed] [Google Scholar]
- 10.Le Hir, H., Gatfield, D., Izaurralde, E. & Moore, M. J. (2001) EMBO J. 20, 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Hir, H., Izaurralde, E., Maquat, L. E. & Moore, M. J. (2000) EMBO J. 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lykke-Andersen, J., Shu, M.-D. & Steitz, J. A. (2001) Science 293, 1836–1839. [DOI] [PubMed] [Google Scholar]
- 13.Lejeune, F., Ishigaki, Y., Li, X. & Maquat, L. E. (2002) EMBO J. 21, 3536–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayeda, A., Badolato, J., Kobayashi, R., Zhang, M. Q., Gardiner, E. M. & Krainer, A. R. (1999) EMBO J. 18, 4560–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCracken, S., Lambermon, M. & Blencowe, B. J. (2002) Mol. Cell. Biol. 22, 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, V. N., Kataoka, N. & Dreyfuss, G. (2001) Science 293, 1832–1836. [DOI] [PubMed] [Google Scholar]
- 17.Le Hir, H., Gatfield, D., Braun, I. C., Forler, D. & Izaurralde, E. (2001) EMBO Rep. 2, 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou, Z., Luo, M.-J., Straesser, K., Katahira, J., Hurt, E. & Reed, R. (2000) Nature 407, 401–405. [DOI] [PubMed] [Google Scholar]
- 19.Rodrigues, J. P., Rode, M., Gatfield, D., Blencowe, B. J., Carmo-Fonseca, M. & Izaurralde, E. (2001) Proc. Natl. Acad. Sci. USA 98, 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cullen, B. R. (2003) J. Cell Sci. 116, 587–597. [DOI] [PubMed] [Google Scholar]
- 21.Gatfield, D. & Izaurralde, E. (2002) J. Cell Biol. 159, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macmorris, M., Brocker, C. & Blumenthal, T. (2003) RNA 9, 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longman, D., Johnstone, I. L. & Cáceres, J. F. (2003) RNA 9, 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Y., Gattoni, R., Stévenin & Steitz, J. A. (2003) Mol. Cell 11, 837–843. [DOI] [PubMed] [Google Scholar]
- 25.Kang, Y. & Cullen, B. R. (1999) Genes Dev. 13, 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai, Z., Gorin, A., Frederick, R., Ye, X., Hu, W., Majumdar, A., Kettani, A. & Patel, D. J. (1998) Nat. Struct. Biol. 5, 203–212. [DOI] [PubMed] [Google Scholar]
- 27.De Gregorio, E., Baron, J., Preiss, T. & Hentze, M. W. (2001) RNA 7, 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, M.-J., Zhou, Z., Magni, K., Christoforides, C., Rappsilber, J., Mann, M. & Reed, R. (2001) Nature 413, 644–647. [DOI] [PubMed] [Google Scholar]
- 29.Hope, T. J., Huang, X., McDonald, D. & Parslow, T. G. (1990) Proc. Natl. Acad. Sci. USA 87, 7787–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald, D., Hope, T. J. & Parslow, T. G. (1992) J. Virol. 66, 7232–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang, D. D. & Sharp, P. A. (1989) Cell 59, 789–795. [DOI] [PubMed] [Google Scholar]
- 32.Wagner, E. & Lykke-Andersen, J. (2002) J. Cell. Sci. 115, 3033–3038. [DOI] [PubMed] [Google Scholar]
- 33.Reed, R. & Hurt, E. (2002) Cell 108, 523–531. [DOI] [PubMed] [Google Scholar]
- 34.Ohno, M., Segref, A., Kuersten, S. & Mattaj, I. W. (2002) Mol. Cell 9, 659–671. [DOI] [PubMed] [Google Scholar]
- 35.Strässer, K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzl, M., Rodriguez-Navarro, S., Rondón, A. G., Agullera, A., Struhl, K., Reed, R. & Hurt, E. (2002) Nature 417, 304–307. [DOI] [PubMed] [Google Scholar]
- 36.Kiesler, E., Miralles, F. & Visa, N. (2002) Curr. Biol. 12, 859–862. [DOI] [PubMed] [Google Scholar]
- 37.Huang, Y., Wimler, K. M. & Carmichael, G. G. (1999) EMBO J. 18, 1642–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]