Abstract
The overall topology of DNA profoundly influences the regulation of transcription and is determined by DNA flexibility as well as the binding of proteins that induce DNA torsion, distortion, and/or looping. Gal repressor (GalR) is thought to repress transcription from the two promoters of the gal operon of Escherichia coli by forming a DNA loop of ≈40 nm of DNA that encompasses the promoters. Associated evidence of a topological regulatory mechanism of the transcription repression is the requirement for a supercoiled DNA template and the histone-like heat unstable nucleoid protein (HU). By using single-molecule manipulations to generate and finely tune tension in DNA molecules, we directly detected GalR/HU-mediated DNA looping and characterized its kinetics, thermodynamics, and supercoiling dependence. The factors required for gal DNA looping in single-molecule experiments (HU, GalR and DNA supercoiling) correspond exactly to those necessary for gal repression observed both in vitro and in vivo. Our single-molecule experiments revealed that negatively supercoiled DNA, under slight tension, denatured to facilitate GalR/HU-mediated DNA loop formation. Such topological intermediates may operate similarly in other multiprotein complexes of transcription, replication, and recombination.
The nucleoid structure in the bacterium Escherichia coli contains a circular DNA molecule of 4.7 million bp present in highly condensed form. The condensation is mediated by DNA supercoiling and the binding of several small nucleoid-associated proteins, e.g., heat unstable nucleoid protein (HU), integration host factor (IHF), factor for inversion stimulation (FIS), histone-like nucleoid structuring protein (HNS), suppressor of thymidylate synthase mutant phenotype A (StpA), and DNA binding protein from starved cells (Dps). These proteins are known to bend DNA or bind to altered structures of DNA. It is suggested that these proteins are mainly responsible for the compaction of DNA in a way that distinguishes the bacterial nucleoid from eukaryotic chromatin. These proteins are also associated with the machinery of macromolecular biosynthesis, including RNA polymerase and specific gene-regulatory, DNA-binding proteins such as repressors and activators. Indeed, DNA may serve as a scaffold for the organized recruitment and assembly of proteins at specific positions to create nucleoprotein complexes with specific activity and regulatory properties. Such positioning has long been postulated to be the mechanism of repression of the gal operon by the gal repressor dimer protein (GalR). GalR represses transcription initiation from the two promoters, P1 and P2, of the gal operon by binding to two spatially separated operators, OE and OI, which encompass the promoters (1). Repression also requires supercoiled DNA and the presence of the nucleoid-associated protein HU (2). It has been proposed that a DNA loop generated by the interaction of the two operator-bound gal repressors inactivates the promoter (3). Repression of the gal operon would, thus, occur when a nucleoprotein complex forms containing supercoiled DNA, two GalR dimers, and HU. However, direct evidence of such looped complex has not been reported previously. Furthermore, an understanding of local conformational changes and their thermodynamic driving forces is required to relate structure to function.
Conventional techniques for studying DNA–protein interactions are restricted to observing the average properties of molecular ensembles. To breach this constraint, we have used magnetic tweezers (4) to unwind and stretch single DNA molecules, containing the regulatory segment of the gal operon, to study the molecular mechanism of transcriptional control by the Gal repressor protein. We reasoned that, if DNA looping was to cause transcriptional repression of the gal operon, it would be possible to detect transitions between short (looped) and long (unlooped) conformations of single, supercoiled gal DNA molecules under moderate tension only in the presence of the GalR and HU proteins (Fig. 1a).
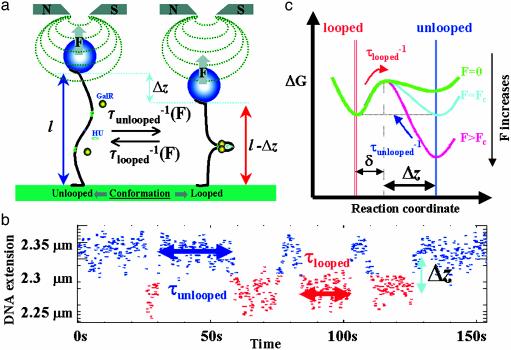
Fig. 1.
Experimental set-up. (a) A single DNA molecule tethering a magnetic bead to a surface can be twisted and stretched by using small magnets placed above the sample. DNA loop formation by GalR and HU reduces the bead-to-surface distance by an amount Δz at the expense of the work, FΔz, performed against the stretching force F. The tension on the DNA may be used to tune the transition rates, (τunlooped)–1 and (τlooped)–1, between the unlooped and looped state. (b) A typical telegraph-like signal. (c) A diagram illustrating the variation of ΔG for the reaction involving the DNA conformational change associated with loop opening. The activation energy for loop opening, Eb – δF, is slightly decreased on pulling, whereas the activation energy for loop formation, Ef + FΔz, is increased on pulling.
Materials and Methods
Sample Preparation and Experimental Set Up. The linear DNA fragments used in the single-molecule experiments are prepared by linearization of plasmid pBR509. pBR509 contains 288 bp of the gal operon (from –197 to 91 bp from transcriptional initiation site) between EcoRI and BamHI restriction sites of pBR322. pBR509 also contains a sequence of 6,497 bp from the digestion of λ phage with BamHI and SphI. Digestion of pBR509 with SalI and EagI allowed ligation of these two ends with two “tails” labeled with biotin and digoxigenin, respectively. The two different DNA “tails” are synthesized by PCR by using biotin-labeled nucleotides in one and digoxigenin-labeled nucleotides in the other. In so doing, the labels will be on both strands of the DNA tail. The biotinated tail and the digoxigeninated one, ≈1000 bp long, will contain an EagI and a SalI site, respectively. In this way, after digestion of the tails, they can be ligated to the opposite ends of the fragment from pBR509. Single molecules of DNA were attached at one end to the glass surface of a microscope flow-chamber and at the other end to a paramagnetic bead 2.8 μm in diameter. Magnetic tweezers (4) were used to twist and pull single DNA molecules attached to the beads, and the length of the DNA was monitored by 3D, video-rate monitoring of the bead (5). By tracking the 3D position of the tethered bead (5, 6), the extension l = 〈z〉 of the molecule can be measured, with an error of ≈10 nm with 1-s averaging. The horizontal motion of the bead 〈Δx2〉 allows the determination of the stretching force via the equipartition theorem: F = kBT l/〈Δx2〉. F was measured with 10% accuracy. To eliminate microscope drift, differential tracking was performed via a second bead glued to the surface.
Data Processing. Traces with transitions between longer (unlooped) to shorter (looped) lengths were best fitted to the raw data l(t) (filtered at 2 Hz) by using a sliding Heaviside (step) function: lstep (t) = sθ(t – t1) + l1 defined over a time window of size Tav. In other words, for every t0 of the data set, the parameters of the step function (s, t1, l1) were fitted such as to minimize the error (l(t) – lstep(t))2 in the time window t0 < t < t0 + Tav, where only one transition is expected. At the end, the parameters that consistently scored best (χ2 test) were selected as steps. The time interval between successive looped and unlooped steps was binned to form histograms of τunlooped (τlooped) corresponding to the time spent in the longer (shorter) state.
Free Energy Calculation: Loop Formation. For an activated process Arrhenius' law yields (7): τunlooped(F) = τ0 exp((Ef + FΔz)/kBT) and τlooped (F) = τ0 exp[(Eb – Fδ)/kBT]. τunlooped and τlooped are the average lifetimes of the unlooped and looped conformations, Ef and Eb are the energy barriers to formation and breakdown of the DNA loop at zero force, kB is Boltzman's constant, T is the temperature, Δz is the decrease in the DNA's extension due to the formation of a loop, and δ is the minimum separation between the two GalR dimers that leads to loop breakdown (6) [about 1 nm (8)]. Considering both looping and unlooping to be first order reactions, the free energy of looping is therefore: ΔGl = kBT ln (τunlooped/τlooped). Notice that at the critical force F = Fc where τunlooped = τlooped: ΔGl = 0. The free energy of looping at zero force is thus: ΔGl,0 = Ef – Eb = –Fc (Δz + δ) (see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org).
Transcription Assays. Supercoiled DNA plasmids (2 nM) were preincubated at 37°C for 5 min in transcription buffer (20 mM Tris acetate/10 mM Mg acetate/50 mM NaCl) supplemented with 1 mM DTT, 1 mM ATP, and 0.8 units recombinant ribonuclease inhibitor (rRNasin) and 20 nM RNA polymerase in a total reaction volume of 50 μl. When required, 80 nM GalR and/or 160 nM HU and/or single-stranded binding protein (SSB) were added before RNA polymerase. To start the transcription reactions, nucleotides were added to a final concentration of 0.1 mM GTP and CTP, 0.01 mM UTP, and 5 μCi [α-32P]UTP (1 Ci = 37 GBq). The reactions were incubated for an additional 10 min before they were terminated by the addition of an equal volume (50 μl) of loading dye (90% formamide/10 mM EDTA/0.1% xylene cyanol/0.1% bromophenol blue). Samples were heated to 90°C for 2–3 min, chilled, then loaded on an 8% sequencing gel and electrophoresed at a constant power of 60 W in TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3). The RNAI transcripts (108 nt) were used as an internal control to quantify the relative number of gal transcripts.
Results and Discussion
DNA molecules that were negatively supercoiled by at least 3% (σ = –0.03) and stretched with a force (F) of 0.88 pN intermittently switched between two conformations in the presence of both GalR and HU (Figs. 1b and 2a). We verified that no telegraphic signals were observed in the absence of GalR and/or HU (data not shown), or in the presence of 40 nM d(+)-galactose, an inducer of gal transcription (Fig. 2a). Similarly, GalR and HU did not generate looping in molecules containing only one operator, OE or OI (data not shown). Taken together, these measurements confirm the widely held hypothesis that GalR and HU induce DNA looping in the gal operon.
Fig. 2.
Traces of DNA length vs. time. The green dots are raw data, whereas the continuous colored lines are the averaged signal. The pertinent conditions are indicated beside each trace. Measurements were conducted at room temperature by using a solution containing 20 mM Tris·HCl (pH 7.8), 1 mM DTT, 50 mM NaCl, and 5 mM MgCl2. The GalR and HU concentrations were 25 nM and 50 nM, respectively. BSA and SSB were used at concentrations of 40 nM. (a) Variations in DNA extension at constant supercoiling (σ =–0.03) and force at F = 0.88 pN, unless otherwise stated. DNA with: no proteins (turquoise), GalR and HU (yellow), GalR and HU with a tension of 1.05 pN (purple), GalR and HU with a tension of 1.32 pN (cyan), GalR and HU and galactose (red), and GalR, HU, and SSB (dark green). (b) DNA extension at F = 0.88 pN, in the presence of GalR and HU as a function of supercoiling. σ = +0.03 (blue); σ = 0 (yellow); σ = –0.015 (purple); σ = –0.03 (cyan); and σ = –0.06 (red). σ is the superhelical density in the DNA defined as (Lk – Lk0)/Lk0, where Lk is the linking number of DNA and is given by the sum of its twist (Tw) and writhe (Wr). In relaxed DNA, Lk = Lk0 = Tw = (number of bp)/10.4 bp. Similarly, in our single-molecule experimental conditions in which DNA molecules are stretched, the distribution between Tw and Wr is about 3:1 whereas in plasmids (unnicked and under no tension) it is about 1:3. As a result, the torsion within each molecule in our measurements at σ = –0.03 (i.e., with Tw ≈ 0.022 Lk0) is slightly higher than the torsion present in a plasmid at sigma = –0.06 (i.e., with Tw ≈ 0.015 Lk0).
The higher force was expected to shift the thermodynamic equilibrium to favor the unlooped form (Fig. 1c). Indeed, the probability of looping decreased when the tension was raised to 1.32 pN (Fig. 2a). Interestingly, the length of DNA that separates the GalR binding sites OE and OI (113 bp or 38 nm for B-DNA) is less than the 55 ± 5 nm transition (Δz) observed experimentally. This finding is consistent with a recently proposed antiparallel loop (9) in which the DNA exiting the loop complex gradually bends to align in the direction of stretching.
Kinetic and Thermodynamic Parameters. By analyzing the lifetimes of the looped and unlooped DNA conformations in traces like those in Fig. 2, we were able to determine the mean lifetimes as a function of force and calculate the free energy changes of the looping reactions. In all cases, the lifetime distributions were well fit by single exponentials (Fig. 3). Thus, in our experiment, loop formation and breakdown can be analyzed as a two-state system. At a force of 0.88 pN, the mean lifetimes for both the looped and unlooped conformations were 17 s. The lifetimes for zero tension were calculated applying the following relations: τunlooped(F) = τunlooped(0) exp(FΔz/kBT), where Δz ≈ 55 nm and τlooped(F) = τlooped(0) exp(–Fδ/kBT), where δ ≈ 1 nm (8) yielding τunlooped(0) ≈ 0.1 ms and τlooped(0) ≈ 21 s. Thus, the lifetime of the unlooped state dramatically increases with the DNA tension, whereas the life time of the GalR/HU-mediated loop remains essentially unchanged. From these lifetimes, we estimated the free energy change involved in the GalR/HU-mediated loop: ΔGl,0 = kBT ln τunlooped(0)/τlooped(0) ≈ –12 kBT or –7.1 kcal/mol (see Materials and Methods, Supporting Text, and refs. 8 and 10). This is a surprisingly large free energy change, commensurate with the 12 kBT liberated by the hydrolysis of one ATP molecule.
Fig. 3.
Mean lifetimes of the looped and the unlooped DNA conformations were calculated from histograms of the lifetimes at F = 0.88 pN and 3% negative supercoiling.
A comparison of this value with that estimated from data reported for the lac repressor protein (LacI) (11), underscores the thermodynamic role, played by the HU protein. In the case of LacI-induced DNA loops, Matthews and colleagues (11) used filter binding of supercoiled plasmids or linear DNA fragments (12) to estimate the unlooping equilibrium constant to be 890 in the presence of supercoiling and 510 times larger in the absence of supercoiling (11). From the relation ΔG = –kBT ln Keq, the free energy for LacI-mediated DNA looping in their supercoiled DNA can be determined to be –6.8 kBT (4 kcal/mol). In single-molecule experiments (13), a similar value of –7 kBT can be calculated from the ratio of mean lifetimes for looped and unlooped states (Keq) of linear DNA in the presence of LacI by using the 510 factor found by Matthews and colleagues (12) to account for supercoiling. Considering that (i) LacI has a higher affinity for its operator site, O1, than GalR has for OE and OI, (ii) LacI is a stable bidentate tetramer, whereas GalR dimers interact weakly, and (iii) the 305-bp lac DNA loop is more flexible than the 113-bp gal DNA loop, HU seems to dramatically stabilize looping in the gal system.
The Role of HU and DNA Supercoiling. To further clarify the roles of HU and DNA supercoiling, we studied DNA loop formation and breakdown in DNA molecules with different values of supercoiling (Fig. 2b) under a condition when the free energy for loop formation is 0 (F ≈ 0.9 pN). Transitions were not observed in either positively supercoiled DNA (σ = +0.03), relaxed DNA (σ = 0), or in DNA unwound by 1.5% (σ = –0.015). However, loop formation and breakdown were observed for DNA unwound by 3% or more (σ = –0.03 or –0.06), which exceeds the 1.5% required to denature AT-rich regions in DNA under tension (14–16). Considering that the HU binding site at the apex of the gal DNA loop is AT rich and palindromic (9, 17), and that HU preferentially binds to altered DNA structures such as denatured or cruciform DNA (18–22), we reasoned that HU may facilitate GalR-mediated looping by tightly binding the single-stranded DNA (ssDNA) in the unwound segment induced by negative supercoiling. Such a conformation would lower the energy required for looping, because ssDNA is more flexible than double-stranded DNA. The model just hypothesized is described in Fig. 4.
Fig. 4.
Suggested model of the mechanism of GalR/HU-induced DNA loop formation. Negative supercoiling favors the opening of a bubble of a few base pairs in the DNA. HU binds preferentially to it, subsequently bending the ssDNA and stabilizing the interaction between two GalR dimers and loop closure.
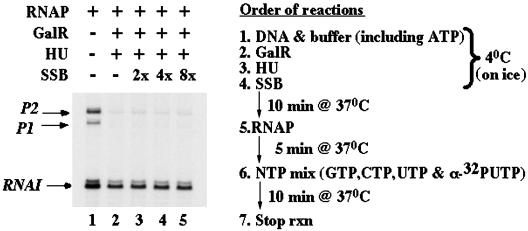
To test this hypothesis, we investigated whether or not SSB interfered with GalR/HU-mediated looping. SSB (23) and HU (24) proteins both have dissociation constants in the nanomolar range for binding to single-stranded DNA. Addition of SSB to our assay eliminated all transitions between looped and unlooped conformations (Fig. 2a), whereas BSA, used as a control, had no effect (data not shown). This result suggested that HU binding to ssDNA might be required for repression of the gal operon. SSB was also used in an in vitro transcription assay using supercoiled plasmids ≈3 kb long. In this case, SSB added before RNA polymerase did not alter simultaneous repression of P1 and P2 by HU and GalR (Fig. 5).
Fig. 5.
In vitro transcription assay. Radiolabeled RNA products of an in vitro transcription assay were separated by gel electrophoresis (Left). A scheme of the procedure is on the Right. SSB added to lanes 3–5 did not interfere with repression of transcription from promoters P1 and P2 due to looping induced by GalR/HU (lane 2). No repression is observed without GalR/HU (lane 1).
Energetic considerations nicely explain the difference between the in vitro transcription and the looping experiments. With 0.9 pN of tension in the DNA, the ΔG of loop formation is 0 and that of SSB binding to DNA is approximately –9 kBT (25). Thus, SSB is an effective competitor of GalR/HU-mediated looping. On the other hand, in the transcription assay in which the DNA is not stretched, the free energy change associated with loop formation becomes approximately –12 kBT. At the same time, SSB binding becomes less favorable given the higher temperature of this assay (26) and cannot compete effectively with the looping reaction. Stretching the DNA deftly shifted the free energy landscape to favor competition by SSB, and it remains to be shown that gal DNA is under similar tension in vivo.
Conclusion
Our work shows that manipulation of single molecules permits the analysis of macromolecular machinery of increasing complexity. Kinetic control achieved through tensioning single DNA molecules permitted the characterization of a loop that is remarkably stabilized thermodynamically by the HU protein. Such a high negative free energy change ensures rapid repression when galactose is unavailable. SSB interfered with the formation of the loop in gently stretched molecules, which might be relevant to conditions in vivo, in which genomic DNA is supercoiled, topologically constrained by proteins, and locally tensioned by DNA processing enzymes (27). Transcriptional control via such a loop would be inherently sensitive to the local environmental and energetic context. The multifactorial and characteristic kinetic and thermodynamic properties of gal DNA looping distinguish a dynamic system for reducing the expression of the products of gal operon (28). Besides induction (loop breakdown) of the operon by d(+)-galactose, which acts by inactivating GalR, the loop must break down for expression in response to anabolic needs resulting from changes in DNA superhelicity or HU concentration (29–31).
Supplementary Material
Acknowledgments
This work was supported by grants from Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) and the Human Frontier Science Program (HFSP) (to L.F.); grants from MIUR (to D.D.); an Erasmus fellowship (to G.L.); grants from Association pour la Recherche sur le Cancer (ARC) (to V.C.), Centre National de la Recherche Scientifique (CNRS), Ecole Normale Supérieure (ENS), Université Paris VI, and Université Paris VII and CEE MolSwitch (to D.B.); support from the French Ministere de la Recherche (to J.-F.A.); and grants from the National Institutes of Health Intramural Research Program (to S.A. and D.E.A.L.).
Abbreviations: HU, heat unstable nucleoid protein; GalR, gal repressor dimer protein; SSB, single-stranded binding protein; ssDNA, single-stranded DNA.
References
- 1.Irani, M. H., Orosz, L. & Adhya, S. (1983) Cell 32, 783–788. [DOI] [PubMed] [Google Scholar]
- 2.Aki, T., Choy, H. E. & Adhya, S. (1996) Genes Cells 1, 179–188. [DOI] [PubMed] [Google Scholar]
- 3.Choy, H. E., Park, S. W., Parrack, P. & Adhya, S. (1995) Proc. Natl. Acad. Sci. USA 92, 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strick, T. R., Allemand, J. F., Bensimon, D. & Croquette, V. (1996) Science 271, 1835–1837. [DOI] [PubMed] [Google Scholar]
- 5.Grosse, C. & Croquette, V. (2002) Biophys. J. 82, 3314–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strick, T. R., Allemand, J. F., Croquette V. & Bensimon, D. (1998) J. Stat. Phys. 93, 647–672. [Google Scholar]
- 7.Rief, M., Fernandez, J. & Gaub, H. (1998) Phys. Rev. Lett. 81, 4764–4767. [Google Scholar]
- 8.Marko, J. F. & Siggia, E. D. (1997) Biophys. J. 73, 2173–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geanacopoulos, M., Vasmetzis, G., Zhurkin, V. B. & Adhya, S. (2001) Nat. Struct. Biol. 8, 432–436. [DOI] [PubMed] [Google Scholar]
- 10.Liphardt, J., Onoa, B., Smith, S. B., Tinoco, I., Jr., & Bustamante, C. (2001) Science 292, 733–737. [DOI] [PubMed] [Google Scholar]
- 11.Whitson, P. A., Hsieh, W.-T., Wells, R. & Matthews, K. S. (1987) J. Biol. Chem. 262, 4943–4946. [PubMed] [Google Scholar]
- 12.Hsieh, W.-T., Whitson, P., Matthews, K. S. & Wells, R. D. (1987) J. Biol. Chem. 262, 14583–14591. [PubMed] [Google Scholar]
- 13.Finzi, L. & Gelles, J. (1995) Science 267, 378–380. [DOI] [PubMed] [Google Scholar]
- 14.Strick, T. R., Croquette, V. & Bensimon, D. (1998) Proc. Natl. Acad. Sci. USA 95, 10578–10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allemand, J.-F., Bensimon, D., Lavery, R. & Croquette, V. (1998) Proc. Natl. Acad. Sci. USA 95, 14152–14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwa, T., Marinari, E., Sneppen, K. & Tang, L. (2003) Proc. Natl. Acad. Sci. USA 100, 4411–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aki, T. & Adhya, S. (1997) EMBO J. 16, 3666–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonnefoy, E., Takahashi, M. & Rouvière-Yaniv, J. (1994) J. Mol. Biol. 242, 116–129. [DOI] [PubMed] [Google Scholar]
- 19.Castaing, B., Zelwer, C., Laval, J. & Boiteux, S. (1995) J. Biol. Chem. 270, 10291–10296. [DOI] [PubMed] [Google Scholar]
- 20.Kobryn, K., Lavoie, D. & Chaconas, G. (1999) J. Mol. Biol. 289, 777–784. [DOI] [PubMed] [Google Scholar]
- 21.Grove, A., Galeone, A., Mayol, L. & Geiduschek, E. P. (1996) J. Mol. Biol. 260, 196–206. [DOI] [PubMed] [Google Scholar]
- 22.Grove, A. & Lim, L. (2001) J. Mol. Biol. 311, 491–502. [DOI] [PubMed] [Google Scholar]
- 23.LeCaptain, D. J., Michel, M. A. & Van Orden, A. (2001) Analyst 126, 1279–1284. [DOI] [PubMed] [Google Scholar]
- 24.Kamashev, D. & Rouvière-Yaniv, R. (2000) EMBO J. 19, 6527–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bujalowski, W. & Lohman T. M. (1987) J. Mol. Biol. 195, 897–907. [DOI] [PubMed] [Google Scholar]
- 26.Overman, L. B., Bujalowski, W. & Lohman, T. (1988) Biochemistry 27, 456–471. [DOI] [PubMed] [Google Scholar]
- 27.Forde, N. R., Izhaky, D., Woodcock, G. R., Wuite, G. J. & Bustamante, C. (2002) Proc. Natl. Acad. Sci. USA 99, 11682–11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adhya, S. (1987) in Escherichia coli and Salmonella typhmirium, ed. Neidhardt, F. (Am. Soc. Microbiol., Washington, DC) pp. 1503–1512.
- 29.Claret, L. & Rouvière-Yaniv, J. (1997) J. Mol. Biol. 273, 93–104. [DOI] [PubMed] [Google Scholar]
- 30.Wei, Y., Lee, J. M., Richmond, C., Blattner, F. R., Rafalski, J. A. & LaRossa, R. A. (2001) J. Bacteriol. 183, 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azam, T. A. & Ishihama, A. (1999) J. Biol. Chem. 274, 33105–33113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.