Abstract
BtuB is a TonB-dependent outer-membrane transporter for vitamin B12 (or cyanocobalamin, CN-Cbl) in Escherichia coli. The binding of CN-Cbl is believed to promote an unfolding or undocking of the Ton box, the conserved N-terminal energy coupling motif at the periplasmic surface of the transporter. This structural change may facilitate the interaction of BtuB with the inner membrane protein TonB. In this work, the effect of the receptor binding fragment of colicin E3 (E3R) on the conformation of the Ton box was examined with site-directed spin labeling. Addition of E3R reverses the undocking of the Ton box that is promoted by CN-Cbl, consistent with a competitive binding between the substrate and the colicin fragment. EPR spectroscopy indicates that the Ton box is in a two-state equilibrium between docked and undocked conformations. In the absence of substrate, the docked conformation is the predominant state; however, the equilibrium can be shifted to the undocked state by the addition of detergents or site-specific proline substitutions. Even when the undocking is induced by detergents or by certain proline mutations, E3R binding shifts the equilibrium to the docked conformation. Thus, two competitive extracellular ligands, CN-Cbl and ER3, transduce opposite conformations of the N-terminal Ton box. Substrate binding stabilizes an undocked conformation, whereas E3R binding stabilizes a docked conformation of the Ton box.
The outer membrane of Gram-negative bacteria such as Escherichia coli contains a family of membrane proteins that actively transport large and scarce nutrients, such as iron siderophores or vitamin B12 (cyanocobalamin, CN-Cbl) into the periplasmic space. These proteins derive energy for transport from the proton chemical potential across the inner membrane by coupling to the transperiplasmic protein TonB and are therefore termed TonB-dependent. High-resolution structures for the vitamin B12 transporter, BtuB, have recently been obtained (1). These structures are similar to other TonB-dependent transporters, such as FepA, FhuA, and FecA (2–5), and consist of a β-barrel formed from 22 antiparallel strands with an N-terminal (core) region that is folded into the lumen of the barrel. However, BtuB, which is the smaller than FepA, FhuA, or FecA, has shorter extracellular loops.
The molecular mechanism of TonB-dependent transport is presently not known, but it likely involves a major structural rearrangement or unfolding of the N-terminal core region of the transporter during the transport cycle (6). Site-directed spin labeling (SDSL) is a technique that is particularly sensitive to the changes in tertiary contact made by a spin-labeled side chain (7), and this method indicates that there are significant conformational changes in the Ton box on CN-Cbl binding. The Ton box is a highly conserved region near the N terminus of the transporter that is believed to participate in interactions between TonB and the TonB-dependent transporter. SDSL indicates that the Ton box segment of BtuB (residues 6–12; see Fig. 1) is docked (or folded) within the barrel of the transporter in the absence of substrate, and that it undocks (or unfolds) and increases its aqueous exposure on addition of CN-Cbl (8, 9). The undocking of the Ton box into the periplasmic space on addition of substrate is consistent with cross-linking results (10, 11), with chemical labeling results presented in an accompanying paper (12), and with substrate-induced structural changes that are seen on the C-terminal side of the Ton box in some crystal structures (4). This substrate-induced structural change is believed to provide a signal at the periplasmic surface that BtuB is loaded with substrate, and it may trigger the interaction between TonB and BtuB and initiate TonB-dependent transport.
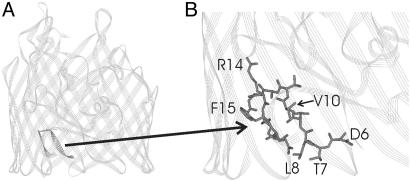
Fig. 1.
Ribbon diagram depicting the crystal structure of the substrate-free form of BtuB (1). (A) Residues 6–17 are highlighted and include the energy-coupling segment or Ton box (residues 6–12). (B) An enlargement with residues 6–17 shown in stick form with several side chains labeled.
In addition to their role in transport, TonB-dependent transporters are exploited as highly specific receptors for colicins. Colicins are large toxin proteins produced by and directed against E. coli strains or related bacteria. In general, the colicin structure consists of three domains that each perform a different function: a central receptor R-domain that mediates binding to an outer-membrane receptor, an N-terminal T-domain that mediates translocation of the colicin across the outer membrane, and a C-terminal domain that elicits lethality by pore forming, nuclease, or other activity (13, 14). Translocation employs one of two pathways. Group A colicins use the tol-dependent system, which includes the proteins TolA, TolB, TolQ, TolR, and Pal, whereas, the group B colicins use the ton-dependent system (13). However, the individual steps and molecular details of either of these pathways are not well understood (see refs. 13 and 15–20 for recent reviews and perspectives). The E colicins belong to group A and target BtuB as their receptor. The receptor domains of colicins E3 and E9 have identical amino acid sequences, but these colicins have different catalytic domains and kill their target cells by different activities. E9 kills its targets by DNA degradation (17), whereas E3 acts as a highly specific RNase (21, 22).
The molecular mechanisms by which colicins interact with TonB-dependent transporters and are translocated are poorly understood (23). In the present work, SDSL and EPR spectroscopy are used to investigate whether the 76-residue receptor binding fragment of colicin E3 (E3R, residues 343–418) induces a conformational change in BtuB. This 76-aa receptor binding fragment has been shown to bind to BtuB with an affinity comparable to that of the intact colicin (23). Here, we determined whether binding of E3R to purified and reconstituted BtuB samples signaled a structural change in the Ton box of BtuB. One hypothesis about translocation is that the type E colicins may pass through BtuB by a “nail” hypothesis (15). Conceivably, E3R might undock the Ton box as a prelude to its transport through BtuB. However, we find that E3R reverses the substrate-induced undocking of the Ton box of BtuB; i.e., the Ton box redocks. Thus, E3R is unlikely to pass although BtuB. More remarkably, E3R is found to stabilize the docked form of the Ton box even when its conformation is undocked by the addition of detergents or by a proline mutation within the Ton box. These results are consistent with data presented in an accompanying study in intact cells (12), in which the maleimide labeling patterns along the Ton box in the presence of E3R were mapped. The EPR spectra provide evidence that in the unliganded state the Ton box exists in an equilibrium between docked and undocked conformations, a result which accounts for the residual maleimide labeling in the absence of substrate. Thus, because E3 and CN-Cbl bind competitively (24), it is not surprising that E3R signals a change in the Ton box conformation opposite to that produced by CN-Cbl, the BtuB substrate.
Methods
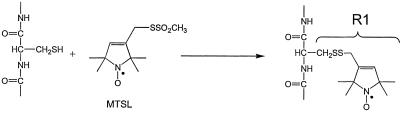
Materials. The spin-labeled sulfhydryl reactive methanethiosulfonate, S-(1-oxy-2,2,5,6-tetramethylpyrroline-3-methyl)methanethiosulfonate (MTSL), was obtained from Toronto Research Chemicals. POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) was purchased from Avanti Polar Lipids, and octyl glucoside (Anagrade) was purchased from Anatrace (Maumee, OH). Bistris [bis(2-hydroxyethyl)imino-tris(hydroxymethyl)-methane] hydrochloride (98%) was purchased from Sigma.
Mutagenesis and Isolation of Intact Outer Membranes. Cysteine mutants were produced by using previously described two-step PCR-based site-directed mutagenesis (10) and were expressed in E. coli in strain RK5016 (metE). This strain is a derivative of strain MC4100 [Δ(argF-lac)U169 araD139 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR22 non-9 gyrA219] with additional mutations (metE70 argH btuB recA), so that vitamin B12 is required for methionine biosynthesis. Outer membranes were isolated as described (25).
Preparation of Colicin E3R. A fragment of colicin E9 corresponding to residues 343–418 has been shown to retain receptor binding activity (23) and has an identical amino acid sequence to the receptor binding domain of colicin E3. This fragment was expressed with a C-terminal His-6 tag and purified as described (12), and will be referred to herein as E3R. A sample of the intact colicin E3 was generously provided by William Cramer (Purdue University, West Lafayette, IN).
Protein Purification, Spin Labeling, and Reconstitution. Outer membranes were solubilized by octyl glucoside, and specific single or double cysteine BtuB mutants were then spin-labeled (Scheme 1) and purified by ion exchange chromatography as described (9, 26). Typically, ≈100 μl of a 22 mM MTSL solution was added to 30 ml of crude detergent-solubilized outer membrane fraction and allowed to react for 2–17 h before purification by anion exchange chromatography on a 5-ml high-performance Sepharose Q prepacked column using an AKTA purifier system (Amersham Biosciences) [buffer A: 25 mM Bistris/16 mM octylglucoside (OG), pH 7.0; buffer B: 25 mM Bistris/16 mM OG/1 M LiCl, pH 7.0]. The purity of BtuB was estimated to be at least 98% as judged by SDS/PAGE electrophoresis. The purified spin-labeled BtuB was reconstituted into POPC vesicles as described (26), by dialysis of mixed micelles having a composition of POPC/OG of 1:17.
Scheme 1.
EPR. EPR spectra were obtained by using a Varian E-line 102 Series X-Band spectrometer fitted with a microwave preamplifier (MITEQ, Hauppauge, NY) and a loop-gap resonator (Medical Advances, Milwaukee, WI). Data collection and analysis were carried out by using labview software generously provided by Christian Altenbach and Wayne Hubbell (University of California, Los Angeles). Samples were placed in glass capillary tubes with a 0.8-mm i.d. (VitroCom, Mountain Lakes, NJ), and EPR spectra were recorded at an incident microwave power of 2.0 mW, a modulation of 1 G peak-to-peak. Spectral subtraction was used to determine the populations of docked and undocked Ton box giving rise to the EPR spectra of spin-labeled BtuB mutants. To carry out the subtraction, the spectra were assumed to arise from two motional components where the narrower spectral component corresponds to the undocked form of the Ton box. The narrower component was simulated by using an isotropic lineshape. There are two major sources of error in the estimate of these populations. First, the spectral subtraction method introduces an error of ≈±5% for the highest percentage values and ranges to <±1% for those spectra that represent the mostly closed state. Second, the true lineshapes of the fully undocked Ton box are not known, and the isotropic lineshape used in the subtraction procedure might not accurately represent this state.
Results
Colicin E3R Reverses the CN-Cbl Substrate-Induced Conformational Change of the Ton Box. SDSL (Scheme 1) is a technique that provides information about local secondary structure and side-chain tertiary contact, as well as conformational changes and backbone dynamics, in proteins (7, 27, 28). The MTSL spin-label was incorporated into sites in the Ton box region of BtuB that were previously shown to exhibit dramatic changes in EPR lineshapes because of loss of tertiary contact and enhanced backbone motion on binding of CN-Cbl (8). Shown in Fig. 2 are EPR spectra of purified and reconstituted BtuB that have a single methanethiosulfonate spin-labeled side chain (R1) at positions 6, 8, and 10 (D6R1, L8R1, and V10R1) and a spectrum of a double-labeled mutant at positions L7R1 and V10R1. The relatively broad spectrum of V10R1 is characteristic of an R1 side chain that is in strong tertiary contact with other portions of the protein, and the spectra for D6R1 and L8R1 are characteristic of R1 attached to a structured protein backbone. As discussed previously, these spectra reflect a configuration for the Ton box that is docked or folded into the BtuB barrel (8, 9). The double-labeled R1 spectrum of T7R1/V10R1 was analyzed as described previously for interacting pairs of nitroxides, and the very weak dipolar broadening found indicates that the spins are separated by a bimodal distance distribution of 12 Å and >18 Å (9, 29).
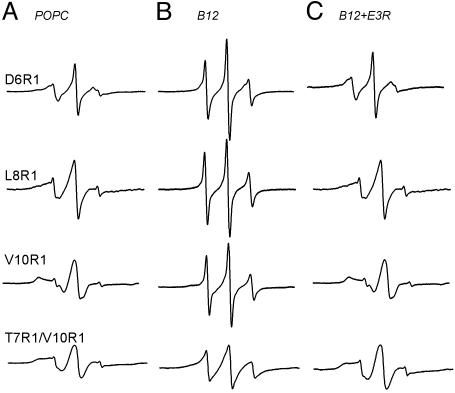
Fig. 2.
X-band EPR spectra of spin-labeled BtuB mutants. (A) Protein reconstituted in POPC (200 μM BtuB). (B) Same protein samples as in A, now in the presence of 300 μM CNCbl. (C) Spectra after addition of 400 μM E3R to the samples in B. All spectra were taken at an incident microwave power of 2 mW, a 1-G modulation amplitude, and a 100-G scan width.
As shown previously, the addition of CN-Cbl changes the conformation of the Ton box, converting it from a form that is docked within the BtuB barrel to an undocked form that has increased mobility and enhanced aqueous exposure (8, 9). The spectra obtained for D6R1, L8R1, V10R1, and T7R1/V10R1 after addition of substrate are shown in Fig. 2B. The narrower lineshapes for the three single-labeled sites in the presence of substrate result from a highly flexible protein backbone that arises from an unfolded form of the Ton box (8). The EPR spectrum of double-labeled T7R1/V10R1 in the undocked state displays features characteristic of labels that are undergoing moderate collisional spin exchange (30). This is not unexpected. In the substrate-bound state, this protein segment is highly flexible, and spin labels attached to the segment would be expected to have a high collision frequency.
Addition of excess E3R to samples already containing vitamin B12 reversed the undocking of the Ton box and resulted in EPR spectra that are identical to those obtained in the absence of substrate (Fig. 2C). These spectra indicate that E3R binding to BtuB refolds the Ton box to its docked conformation after it has been unfolded by substrate. If these samples are subjected to subsequent additions of excess CN-Cbl followed by excess E3R, the Ton box will continue to cycle between undocked and docked configurations, respectively.
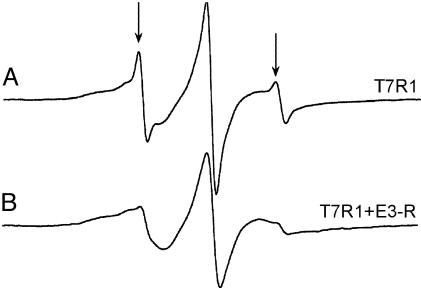
Colicin E3R Stabilizes the Docked Configuration of the Ton Box. The observation that E3R reverses the effect of CN-Cbl is not entirely unexpected. Because these two ligands compete for sites on the external surface of BtuB, E3R should displace bound substrate. Removal of CN-Cbl might restore the native form of the Ton box. However, E3R also appears to independently stabilize the docked state of the Ton box. This point is demonstrated by the following three experiments. First, when BtuB samples are reconstituted into POPC vesicles, some R1-labeled mutants yield EPR lineshapes that have easily distinguishable narrower components. For example, Fig. 3 shows the EPR spectrum of T7R1 in POPC vesicles. This spectrum is a mixture of two motional components, where the minor narrower component represents <10% of the total spin signal. The lineshape of this narrower component is identical to the lineshape of T7R1 after addition of substrate and is believed to arise from a population of the Ton box in its undocked conformation. From the relative intensities of both populations, it is estimated that the docked and undocked conformations are close in energy (for this R1 mutant, these conformations are estimated to differ by ≈1.3 kcal/mol). The fact that both populations are observable indicates that the conversion between these forms is slow on the EPR time scale (nanoseconds). When E3R is added in excess to this reconstituted BtuB mutant, the narrow component in the EPR spectrum is virtually eliminated (Fig. 3), thus suggesting that E3R stabilizes the docked conformation of the Ton box of BtuB.
Fig. 3.
X-band EPR spectra of T7R1 in POPC (A) and with E3R added (B). The arrows indicate the positions of the narrower lineshape component that appears in several spectra in the presence of POPC. This component represents a minor population of the Ton box that is present in an undocked configuration in these POPC membranes. The fraction of undocked Ton box varies with the position of the R1 substitution and with the lipid composition. Spectra were acquired with an incident microwave power of 2 mW, a 1-G modulation amplitude, and a 100-G scan width.
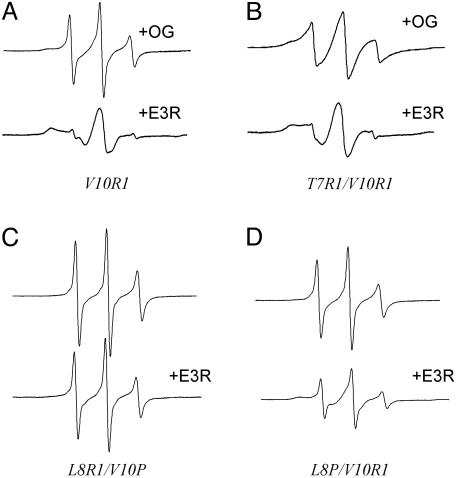
More dramatic evidence for this stabilization is obtained from experiments where the effect of E3R on BtuB solubilized in mixed micelles of detergent and lipid is investigated. The addition of OG or decylmaltoside (DM) to form mixed micelles of BtuB/POPC/detergent unfolds the N terminus of BtuB including the Ton box. Shown in Fig. 4 A and B are the EPR spectra of V10R1 and T7R1/V10R1 in the presence of mixed micelles of OG/POPC (17:1). These spectra resemble those obtained for these mutants in the presence of CN-Cbl. Remarkably, the addition of E3R to this protein-mixed micelle system restores the Ton box to its docked state as indicated by the restoration of the native-like EPR spectra for V10R1 and L7R1/V10R1 (compare with Fig. 2 A). These results provide a clear indication that the undocked configuration of the Ton box, induced by the addition of detergent, can be reversed by the addition of colicin E3R.
Fig. 4.
(A) Comparison of the EPR spectra of spin-labeled BtuB mutants (≈200 μM) of V10R1 in mixed micelles of 17:1 OG:POPC (Upper) and with 400 μM E3R added (Lower). (B) T7R1/V10R1 in mixed micelles of 17:1 OG/POPC (Upper) and with 400 μM E3R added (Lower). (C) L8R1/V10P in POPC (Upper) and with 200 μM E3R added (Lower). (D) L8P/V10R1 in POPC (Upper) and with 200 μM E3R added (Lower). The incomplete closure of L8P/V10R1 in D results because E3R was not added in large molar excess.
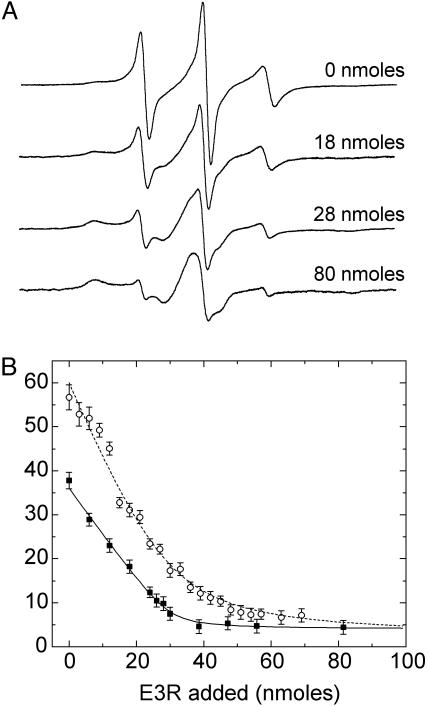
Shown in Fig. 5A are a series of EPR spectra for the BtuB mutant V10R1 in mixed micelles showing the lineshape changes that occur when E3R is titrated into the sample. Addition of colicin E3R results in the conversion of the EPR spectrum from a lineshape having relatively narrow features superimposed on a broad underlying component to one that is now dominated by the broad component with only a fraction of the narrow features detectable. Throughout the titration, each of these spectra contains two components, and titration of the sample with colicin E3R alters the relative populations of these spectral components. For a series of these spectra, the relative fraction of the two components has been determined by spectral subtraction, and the fraction of spins found in the undocked state is plotted in Fig. 5B. Also plotted is the fit to a simple 1:1 binding model (solid line in Fig. 5B). This model assumes that the Ton box has two states and that binding of E3R to BtuB places the Ton box in its docked configuration. The fit provides further evidence that there are two states of the Ton box revealed in the EPR spectra, and that E3R stabilizes the folded or docked state. Also included in Fig. 5B are the titration results for E3R to a V10R1 sample that had an equal molar amount of CN-Cbl at the starting point.
Fig. 5.
(A) X-band EPR spectra showing the titration of the spin-labeled mutant V10R1 in mixed micelles with 0, 18, 28, and 80 nmol of E3R. The spectra are 100-G scans. (B) Filled squares represent the fraction of the Ton box in the undocked state as a function of the mol percentage of E3R. The open circles represent the percentage in the undocked state for the V10R1 sample that had an equal molar amount of vitamin B12 added at the beginning of the titration. The percentage of the undocked Ton box was estimated by spectral subtraction and assumes that there are only two motional components giving rise to the EPR spectrum. The solid line represents a fit to the data assuming a 1:1 binding of E3R to BtuB. The apparent 1:1 association constant was estimated to be at least 2 × 106 or greater, but it is not well determined because of the high protein and E3R concentrations used in this titration. The data obtained in the presence of vitamin B12 were also fit assuming a 1:1 binding (dashed line), but this simple model does not account for the competition occurring between the two ligands. The titration experiments were performed on an ≈100 μM BtuB sample in a mixed micelle environment with volumes of 100 μM E3R solution added stepwise.
A final demonstration that E3R binding stabilizes the docked conformation of the Ton box comes from studies where perturbations of the Ton box conformation are induced by amino acid substitutions. Previous SDSL studies have shown that proline mutants L8P or V10P, which result in a transport-defective phenotype, result in an undocked conformation of the Ton box (25). The EPR spectra of both L8P/V10R1 and L8R1/V10P in the absence of substrate have lineshapes characteristic of an unfolded protein backbone (Fig. 4 C and D). Addition of E3R to these samples resulted in the docked conformation of the Ton box for the mutant L8P/V10R1 but not for the L8R1/V10P mutant. Hence, E3R has the capacity to stabilize the docked configuration of the Ton box when the equilibrium is shifted toward the undocked state by an unfavorable amino acid substitution. Apparently, the proline at position 8 is less perturbing than the proline at position 10, and we speculate that E3R addition cannot overcome the energetics of the proline substitution at position 10. Taken together, these three experiments provide strong evidence that the Ton box conformation exists in equilibrium between a docked and undocked state, and that E3R binding shifts this equilibrium toward the docked state. Not only does E3R binding reverse the conformational change induced by CN-Cbl binding, it also transduces a signal through BtuB that stabilizes the docked rather than the undocked state of the Ton box.
It should be noted that in addition to the colicin receptor fragment, E3R, we tested the ability of the intact colicin E3 to redock the Ton box after the addition of substrate (BtuB in POPC) or after detergent-induced unfolding of the Ton box. In either case, the Ton box was refolded by E3 and the behavior appears to be identical to that of the receptor fragment (data not shown).
Discussion
In the work described here, EPR spectroscopy was used to investigate the effect of colicin E3R binding on the conformation of the Ton box of the vitamin B12 transporter BtuB. Previous work from our laboratories provided evidence from chemical cross-linking and SDSL that the binding of substrate altered the conformation of the Ton box of BtuB so that it converted from a form that was docked into the BtuB barrel to a form that was undocked, more dynamic, and had increased aqueous exposure (8, 10, 11). The data presented here provide strong evidence that the binding of a receptor fragment of colicin E3 also alters the conformation of the Ton box of BtuB. Remarkably, the data indicate that colicin binding stabilizes the docked configuration of the Ton box and thus has an effect on the Ton box conformation that is opposite to that of CN-Cbl. The E3R fragment used here is identical in sequence to the binding domain of E9 and similar in sequence to colicins E2, E6, E7, and E8 (17). Thus, the binding of these colicins may elicit the same conformational change of the Ton box as seen with E3 and E3R.
Colicin lethality is a complex, highly coordinated process. To kill the target cell, colicins must translocate the outer and inner membranes, and the details of the process are not understood. After binding to BtuB, it is thought that the N-terminal domain of the E-type colicins unfolds and passes through another outer membrane protein. In the case of colicin E3/E9, OmpF has been shown to be required for translocation of the N terminus (13) and has recently been shown to enhance the ability of BtuB to protect cells from infection with E9 (31). After reaching the periplasm, the N-terminal domain then interacts with the Tol proteins, thereby resulting in outer membrane fragility (32). These events then allow the C-terminal domain to enter into the periplasmic space, but the mechanism of entry into the periplasm is still unknown. Recent work indicates that the C-terminal fragment of colicin E9 has the capacity to form pores (33), and it seems likely that this and other catalytic colicins may initiate their own translocation across the inner membrane once inside the periplasm. The results presented here are consistent with this mechanism for binding and translocation and suggest that colicin E3 is unlikely to pass through the BtuB barrel, thus requiring some other component in the translocation process. Our results show that colicin binding stabilizes the docked conformation of the Ton box. This stabilization may ensure that there is no interaction between BtuB and TonB, and that there is no opening of the transport pore or progression of BtuB through its transport cycle.
The EPR data presented here are consistent with the results presented in the accompanying paper (12), which shows that site-directed cysteines along the Ton box have limited reactivity toward a sulfhydryl-specific biotin maleimide [1-biotinamido-4-[4′-(maleimidomethyl)-cyclohexane-carboxamido]-butane (BMCC)] in the absence of substrate. BMCC results indicate that the Ton box is partially protected, consistent with a docked conformation in the absence of substrate. Addition of substrate increases the reactivity of cysteines along the Ton box segment to BMCC, demonstrating that CN-Cbl increases the aqueous exposure of the Ton box and consistent with an undocking of the Ton box inferred from the EPR data. In this BMCC labeling study, addition of E3R in the absence of substrate results in a more limited labeling pattern of derivatization along the Ton box. This could represent a third, more protected configuration for the Ton box. However, the EPR data presented here strongly suggest that the Ton box is in equilibrium between docked and undocked states, and the BMCC labeling in the presence of E3R data can be easily be interpreted in terms of this equilibrium. The minimal labeling reactivity along the Ton box that is observed in the BMCC study in the absence of substrate likely takes place because the Ton box is in exchange between docked (less reactive) and undocked (more reactive) configurations. The lowered reactivity of BMCC along the Ton box in the presence of E3R likely occurs because E3R binding shifts the equilibrium further toward the docked (less reactive) state of this segment.
The BMCC labeling (12) and EPR studies carried out here and elsewhere (8, 9) provide strong evidence that CN-Cbl binding converts the Ton box from a form that is docked or folded within the BtuB barrel to a form that has greater aqueous exposure. The enhanced backbone dynamics seen by EPR on CN-Cbl addition are characteristic of an unfolded protein segment. Although the high-resolution crystal structure shows evidence for a substrate-induced conformational change (1), this change is minor compared with the changes inferred from BMCC label reactivity or EPR. For example, the EPR data show evidence for a loss of side-chain tertiary contact at all positions along the Ton box, whereas these contacts are not lost on CN-Cbl binding in the crystal structure. There are clearly many experimental differences that might account for the differences observed between labeling or EPR measurements and protein crystallography. However, some of our recent work indicates that the environment of the membrane protein can affect the energetics of the undocking transition. Specifically, we find that solutions with high osmolyte concentrations [having osmotic pressures similar to the solutions used for BtuB crystallography (34)] alter the Ton box conformation and can even block the substrate-induced undocking of the Ton box (G.E.F., J. Y. Lee, and D.S.C., unpublished work). Solutes are well documented to modulate ion channels and water-soluble proteins indirectly through osmotic pressure effects and preferential hydration (35–37), and more work is presently being performed within our laboratories to investigate these effects on BtuB.
In summary, the Ton box of BtuB can exist in either a docked or undocked configuration. When the transporter binds substrate, the Ton box adopts an undocked configuration. However, when the receptor fragment of colicin E3 is bound to BtuB, the docked configuration of the Ton box is stabilized. Both colicin E3 and CN-Cbl interact with the extracellular loops of BtuB and bind competitively. Remarkably, the work presented here indicates that substrate and E3R induce opposite effects on the energetics of the Ton box segment of BtuB. A particularly interesting structural challenge will be to determine how these two extracellular ligands transduce opposite conformations to the N-terminal Ton box on the periplasmic protein surface.
Acknowledgments
We thank Professor William A. Cramer for generously providing a sample of the intact colicin E3 used in this work. This work was supported by National Institutes of Health Grants GM35215 (to D.S.C.) and GM19078 (to R.J.K.) and by National Institutes of Health National Research Service Award Postdoctoral Fellowship GM20298 (to G.E.F.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations:BMCC,1-biotinamido-4-[4′-(maleimidomethyl)-cyclohexane-carboxamido]-butane; CN-Cbl, cyanocobalamin; E3R, 76-aa receptor binding fragment of colicin E3; OG, octylglucoside;POPC,1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine;R1,methanethiosulfonate spin-labeled side chain; SDSL, site-directed spin labeling.
References
- 1.Chimento, D. P., Mohanty, A. K., Kadner, R. J. & Wiener, M. C. (2003) Nat. Struct. Biol. 10, 394–401. [DOI] [PubMed] [Google Scholar]
- 2.Buchanan, S. K., Smith, B. S., Venkatramanil, L., Xia, D., Esser, L., Palnitkar, M., Chakraborty, R., van der Helm, D. & Deisenhofer, J. (1999) Nat. Struct. Biol. 6, 56–63. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson, A. D., Hofmann, E., Coulton, J. W., Diederichs, K. & Welte, W. (1998) Science 282, 2215–2220. [DOI] [PubMed] [Google Scholar]
- 4.Locher, K. P., Rees, B., Koebnik, R., Mitschler, A., Moulinier, L., Rosenbusch, J. P. & Moras, D. (1998) Cell 95, 771–778. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson, A. D., Chakraborty, R., Smith, B. S., Esser, L., van der Helm, D. & Deisenhofer, J. (2002) Science 295, 1715–1719. [DOI] [PubMed] [Google Scholar]
- 6.Usher, K. C., Ozkan, E., Gardner, K. H. & Deisenhofer, J. (2001) Proc. Natl. Acad. Sci. USA 98, 10676–10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbell, W. L., Cafiso, D. S. & Altenbach, C. A. (2000) Nat. Struct. Biol. 7, 735–739. [DOI] [PubMed] [Google Scholar]
- 8.Merianos, H. J., Cadieux, N., Lin, C. H., Kadner, R. & Cafiso, D. S. (2000) Nat. Struct. Biol. 7, 205–209. [DOI] [PubMed] [Google Scholar]
- 9.Fanucci, G. E., Coggshall, K. A., Cadieux, N., Kim, M., Kadner, R. J. & Cafiso, D. S. (2003) Biochemistry 42, 1391–1400. [DOI] [PubMed] [Google Scholar]
- 10.Cadieux, N. & Kadner, R. J. (1999) Proc. Natl. Acad. Sci. USA 96, 10673–10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cadieux, N., Bradbeer, C. & Kadner, R. (2000) J. Bacteriol. 182, 5954–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadieux, N., Phan, P. G., Cafiso, D. S. & Kadner, R. J. (2003) Proc. Natl. Acad. Sci. USA 100, 10688–10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazdunski, C. J., Bouveret, E., Rigal, A., Journet, L., Lloubes, R. & Benedetti, H. (1998) J. Bacteriol. 180, 4993–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schendel, S. L., Click, E. M., Webster, R. E. & Cramer, W. A. (1997) J. Bacteriol. 179, 3683–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao, Z. & Klebba, P. E. (2002) Biochimie 84, 399–412. [DOI] [PubMed] [Google Scholar]
- 16.Zakharov, S. D. & Cramer, W. A. (2002) Biochim. Biophys. Acta 1565, 333–346. [DOI] [PubMed] [Google Scholar]
- 17.James, R., Penfold, C. N., Moore, G. R. & Kleanthous, C. (2002) Biochimie 84, 381–389. [DOI] [PubMed] [Google Scholar]
- 18.Collins, E. S., Whittaker, S. B., Tozawa, K., MacDonald, C., Boetzel, R., Penfold, C. N., Reilly, A., Clayden, N. J., Osborne, M. J., Hemmings, A. M., et al. (2002) J. Mol. Biol. 318, 787–804. [DOI] [PubMed] [Google Scholar]
- 19.Anderluh, G., Hong, Q., Boetzel, R., MacDonald, C., Moore, G. R., Virden, R. & Lakey, J. H. (2003) J. Biol. Chem. 278, 21860–21868. [DOI] [PubMed] [Google Scholar]
- 20.Braun, V., Patzer, S. I. & Hantke, K. (2002) Biochimie 84, 365–380. [DOI] [PubMed] [Google Scholar]
- 21.Zarivach, R., Ben-Zeev, E., Wu, N., Auerbach, T., Bashan, A., Jakes, K., Dickman, K., Kosmidis, A., Schluenzen, F., Yonath, A., et al. (2002) Biochimie 84, 447–454. [DOI] [PubMed] [Google Scholar]
- 22.Soelaiman, S., Jakes, K., Wu, N., Li, C. & Shoham, M. (2001) Mol. Cell 8, 1053–1062. [DOI] [PubMed] [Google Scholar]
- 23.Penfold, C. N., Garinot-Schneider, C., Hemmings, A. M., Moore, G. R., Kleanthous, C. & James, R. (2000) Mol. Microbiol. 38, 639–649. [DOI] [PubMed] [Google Scholar]
- 24.Cavard, D. (1994) FEMS Microbiol. Lett. 116, 37–42. [DOI] [PubMed] [Google Scholar]
- 25.Coggshall, K. A., Cadieux, N., Piedmont, C., Kadner, R. & Cafiso, D. S. (2001) Biochemistry 40, 13946–13971. [DOI] [PubMed] [Google Scholar]
- 26.Fanucci, G. E., Cadieux, N., Piedmont, C. A., Kadner, R. J. & Cafiso, D. S. (2002) Biochemistry 41, 11543–11551. [DOI] [PubMed] [Google Scholar]
- 27.Hubbell, W. L., Gross, A., Langen, R. & Lietzow, M. A. (1998) Curr. Opin. Struct. Biol. 8, 649–656. [DOI] [PubMed] [Google Scholar]
- 28.Columbus, L. & Hubbell, W. L. (2002) Trends Biochem. Sci. 27, 288–295. [DOI] [PubMed] [Google Scholar]
- 29.Altenbach, C., Cai, K., Klein-Seetharaman, J., Khorana, H. G. & Hubbell, W. L. (2001) Biochemistry 40, 15471–15482. [DOI] [PubMed] [Google Scholar]
- 30.Wertz, J. E. & Bolton, J. R. (1972) Electron Spin Resonance: Elementary Theory and Practical Applications (McGraw–Hill, New York).
- 31.Law, C. J., Penfold, C. N., Walker, D. C., Moore, G. R., James, R. & Kleanthous, C. (2003) FEBS Lett. 545, 127–132. [DOI] [PubMed] [Google Scholar]
- 32.Lazzaroni, J. C., Dubuisson, J. F. & Vianney, A. (2002) Biochimie 84, 391–397. [DOI] [PubMed] [Google Scholar]
- 33.Mosbahi, K., Lemaitre, C., Keeble, A. H., Mobasheri, H., Morel, B., James, R., Moore, G. R., Lea, E. J. A. & Kleanthous, C. (2002) Nat. Struct. Biol. 9, 476–484. [DOI] [PubMed] [Google Scholar]
- 34.Chimento, D. P., Mohanty, A. K., Kadner, R. J. & Wiener, M. C. (2003) Acta Crystallogr. D 59, 509–511. [DOI] [PubMed] [Google Scholar]
- 35.Vodyanoy, I., Bezrukov, S. M. & Parsegian, V. A. (1993) Biophys. J. 65, 2097–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colombo, M. F., Rau, D. C. & Parsegian, V. A. (1992) Science 256, 1335–1336. [DOI] [PubMed] [Google Scholar]
- 37.Timasheff, S. N. (1993) Annu. Rev. Biophys. Biomol. Struct. 22, 67–97. [DOI] [PubMed] [Google Scholar]