Abstract
In genetic hybrids, nucleolus formation on chromosomes inherited from only one parent is the epigenetic phenomenon, nucleolar dominance. By using Arabidopsis suecica, the allotetraploid hybrid of Arabidopsis thaliana and Arabidopsis arenosa, natural variation in nucleolar dominance was found to occur, providing a unique opportunity to examine homologous nucleolus organizer regions (NORs) in their active and inactive states. In A. suecica strain LC1, NORs derived from A. arenosa are active, whereas A. thaliana-derived NORs are silenced. In A. suecica strain 9502, NORs of both parental species are active. When active, NORs are partially, but not fully, decondensed. Both active and inactive LC1 NORs colocalize with the nucleolus, contradicting the long-standing assumption that rRNA gene transcription drives nucleolus association. Collectively, these observations clarify the relationships among NOR chromatin topology, rRNA gene transcription, and NOR–nucleolus associations. A. suecica strains LC1 and 9502 have each lost one pair of A. thaliana NORs during evolution, and amplified fragment-length polymorphism analysis further indicates that these strains are genetically very similar. These data suggest that nucleolar dominance can result from subtle genetic or epigenetic variation but is not a trait fundamental to a given interspecies hybrid combination.
In genetic hybrids or allopolyploids, nucleoli often assemble at specific chromosomal loci of one parent but not the other. This phenomenon, known as nucleolar dominance (1–4), was initially discovered as a change in chromosome structure (5). At nucleolus organizer regions (NORs), the loci where nucleoli form during interphase (6, 7), and where genes encoding the precursor transcript for 18S, 5.8S, and 25S rRNA are tandemly arrayed (8–10), NOR-bearing chromosomes in pure species (nonhybrids) display thin “secondary constrictions” at metaphase (6, 7). Navashin noted that in numerous interspecies hybrids, only the chromosomes of one parent display these secondary constrictions (5). Nuclear run-on assays later suggested that transcription of only one parental set of rRNA genes is the molecular explanation for nucleolar dominance (11, 12). Presumably, transcription of rRNA genes during interphase somehow reduces their condensation at metaphase, thus explaining the appearance of secondary constrictions (13).
A recurrent feature of nucleolar dominance in both plants and animals is that the NORs of the same species are always silenced, independent of maternal or paternal effects. This finding suggests that fundamental differences dictate the dominant and repressed sets of rRNA genes in a hybrid, possibly because of species-specific differences in the rRNA gene–RNA polymerase I transcription systems of the progenitors (1, 14). In keeping with the expectation of silencing in only one direction, Arabidopsis arenosa-derived rRNA genes were shown to be transcriptionally dominant over Arabidopsis thaliana-derived rRNA genes, both in a natural Arabidopsis suecica strain and in newly formed A. suecica-like allotetraploid hybrids (15). However, we show here that nucleolar dominance is not a fundamental property of A. suecica. Instead, A. thaliana-derived NORs can be either silenced or active in different natural strains of A. suecica. This discovery of natural variation in nucleolar dominance presented a unique opportunity to study the same NORs in an active or silenced state, an opportunity we exploited to deduce the relationships between NOR chromatin topology, transcriptional activity, and NOR localization relative to the nucleolus. We show that partial, but not complete, NOR decondensation is the cytogenetic manifestation of active rRNA gene transcription. Both active and inactive NORs colocalize with the nucleolus, indicating that nucleolar association is not a reliable indicator of rRNA gene activity. Our analyses also reveal the number of NORs in A. arenosa and A. suecica. We find that one pair of A. thaliana NORs has been lost in A. suecica strains that differ with respect to A. thaliana NOR silencing, and the same pair is lost in both, indicating that NOR loss does not explain the variation in NOR activity. Amplified fragment-length polymorphism (AFLP) analyses further indicate that the strains are genetically similar. Collectively, these studies indicate that nucleolar dominance is not a fundamental trait of A. suecica due to species-specific differences inherent in its progenitors, but it is likely to be dictated by more subtle allelic or epigenetic differences.
Materials and Methods
Plant Material. A. suecica strain LC1 is a laboratory strain derived from Sue-1, provided by L. Comai (16) and reportedly the same strain examined by Hanfstingl et al. (17). The LC1 strain has undergone several generations of single-seed descent in the Pikaard laboratory but is likely to be identical with Sue-1. The original wild population from which Sue-1 and LC1 are derived is unclear, although all known populations of A. suecica are restricted to northern Europe. A. suecica laboratory strain 9502 was derived from a plant of accession 90-10-085-10 (originating in Finland). Laboratory strain 94-53 was derived from a plant of accession 94-53-30-94-00 (collected in a botanical garden in Goettingen, Germany; the original location of the wild population is unknown). Plants of accessions 90-10-085-10 and 94-53-30-94-00 were made available by Steve O'Kane, as was A. arenosa accession 3651 (originating in Poland) (18). A. thaliana ecotype No-0 (Nossen, originating in Germany) was obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Plants used for cytogenetic examinations were grown in a greenhouse (20-h photoperiod at 25 ± 2°C). Roots for cytological preparations were fixed in ethanol/acetic acid (3:1 vol/vol) and stored at –20°C until use. Interphase nuclei and chromosome spreads were prepared according to ref. 19.
Fluorescence in Situ Hybridization (FISH) Probe Labeling. DNA clones used for FISH analyses were pARR20-1, containing a 180-bp A. thaliana-specific pericentromeric repeat (20); pAt2, which corresponds to A. thaliana rRNA gene intergenic spacer (IGS) sequences from –2590 to +92 relative to the transcription start site, +1 (21); and pCaIGS, a complete A. arenosa rRNA gene IGS that was amplified by PCR with 25S and 18S rRNA-coding sequence primers flanking the IGS (D. A. Hayworth and B. A. Schaal, GenBank accession no. AF177417). The rRNA gene probes were labeled with biotin-dUTP or digoxigenin-dUTP by using a nick translation kit and conditions recommended by the supplier (Roche Applied Science). The pericentromeric repeat probe was amplified from pARR20-1 by PCR with the primers 5′-ATCCTCTAGAGTCGACCTGCA-3′ and 5′-TTCCCAGTCACGACGTTGTAA-3′, an initial denaturation step for 4 min at 94°C, and 35 cycles of 94°C for 45 s, 56°C for 45 s, and 72°C for 45 s. The resulting 180-bp PCR products were labeled with digoxigenin-dUTP.
FISH. FISH in cell spreads was performed according to Jones and Heslop-Harrison (19). The hybridization mixture contained 100–200 ng of each probe in 50% formamide/2× SSC/10% dextran sulfate/salmon sperm blocking DNA (10 μg/μl)/0.1% SDS. The mixture was heated for 10 min at 70°C and then incubated on ice for a minimum of 5 min. The chromosome preparations and hybridization mixture were then denatured at 75°C for 10 min on a hot plate. Hybridization was carried out overnight in a moist chamber at 37°C. Two posthybridization washes, 5 min each, were performed in 50% formamide/0.1× SSC at 42°C. Digoxigenin-labeled probes were detected with anti-digoxygenin-fluorescein (Roche Applied Science) and biotin-labeled probes with Cy3-streptavidin (Sigma) according to Jones and Heslop-Harrison (19). Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole hydrochloride (DAPI) in Citifluor antifade buffer (AF1; Agar Scientific, Stansted, U.K.). Epifluorescence microscopy (Zeiss Axioskop 2) images were obtained by using a Zeiss AxioCam digital camera. Confocal FISH analyses used root-tips prepared according to ref. 34, propidium iodide counterstaining, and imaging with a MRC-600 confocal scanning laser microscope (Bio-Rad). Digital images were composed by using photoshop (Adobe Systems, Mountain View, CA).
DNA Isolation. Nucleic acids of A. suecica laboratory strains LC1, 9502, and 94-53 were isolated from leaf tissue frozen in liquid nitrogen, ground to a powder, and mixed vigorously with 3 vol (wt/vol) of extraction buffer (250 mM Tris·HCl, pH 8.5/375 mM NaCl/25 mM EDTA/1% SDS/1% 2-mercaptoethanol/0.5 mg/ml heparin) in a 15-ml snap-cap tube. The resulting homogenate was subjected to centrifugation at 3,000 × g for 10 min to pellet insoluble debris. The aqueous phase was extracted twice with phenol/chloroform, and total nucleic acids were precipitated by addition of 2 vol of ethanol. After centrifugation, pellets were resuspended in sterile water and large RNAs were precipitated with 3 M LiCl. Genomic DNA in the supernatant was recovered by ethanol precipitation and purified further by using a Geneclean Turbo Kit (BIO 101 Systems, Qbiogene, Carlsbad, CA).
RNA Isolation and S1 Nuclease Protection Assay. Twenty micrograms of LiCl-precipitated RNA was hybridized with oligonucleotide probes corresponding to the non-RNA (antisense) strand of A. thaliana or A. arenosa rRNA genes. Probes spanned the transcription start sites and were 5′ end-labeled by using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. The sequence of the A. thaliana-specific probe was 5′-GGGT TCCCCACGGACTGCCCAGACTCCCTCA ACACCCACCCCCCTATATAGCTGCC-3′; the A. arenosa-specific probe was 5′-GGAACCGAGTAGGGAGGTACCCTCGGTCTGCCCAGACTTCACCAACACCCACCCCCTATATAGCTTTTT-3′. After an initial denaturation step at 99°C for 15 min, probe-RNA hybridization reactions were incubated overnight at 50°C. Probe-RNA hybrids were then subjected to S1 nuclease (Invitrogen) digestion (750 units/ml) at 50°C for 45 min. Resulting digestion products were resolved on a urea/10% polyacrylamide sequencing gel. Gels were dried onto filter paper and exposed to x-ray film.
AFLP Analysis. AFLP assessment was conducted by using the AFLP Analysis System II with adapters, primers, and protocols provided by the supplier (GIBCO). In brief, 250 ng of purified genomic DNA was digested to completion by using the restriction endonucleases EcoRI and MseI. Digested DNAs were then ligated to EcoRI and MseI adapters, and the resulting ligation products were amplified by PCR with primers matching the adapters. Resulting PCR products provided the templates for subsequent PCR that reduced the complexity of the DNA fragment pool by using selective primers. These latter selective PCR used an EcoRI-AC primer (EcoRI adapter primer extended on the 3′ end with an adenosine and a cytosine), 5′ end-labeled by using T4 polynucleotide kinase and [γ-32]ATP, together with one of several primers specific to the MseI end of the fragments (M-CAA, M-CAC, M-CAT, M-CTC, M-CTA, and M-CTT). Each primer pair amplifies a distinct subset of DNA fragments in the template mix. Resulting PCR fragments were resolved on a 7.5 M urea/6% polyacrylamide (19:1 acrylamide/bisacrylamide) sequencing gel. The gel was vacuum dried onto filter paper and exposed to x-ray film for 24–48 h.
Results
Origins of NORs in A. suecica. Although A. thaliana NORs have been well characterized cytogenetically, physically, and genetically in the ecotypes Columbia and Landsberg erecta (23–31), the numbers and locations of NORs in A. arenosa, A. suecica, and most ecotypes of A. thaliana are unknown. Using A. thaliana and A. arenosa rRNA gene IGS sequences as FISH probes, we examined the chromosomes of A. suecica and its progenitors (Figs. 1 and 2). The genomes of A. suecica strains LC1 and 9502 include a transposable element found in A. thaliana ecotype No-0 but missing from most ecotypes of A. thaliana, suggesting that No-0 may be related to one progenitor of LC1 and 9502 (32). In A. thaliana ecotype No-0, four NOR FISH signals are observed among the 10 chromosomes in diploid root-tip cells (Fig. 1 A and B). These signals are presumed to correspond to NOR2 and NOR4 (two of each NOR in a diploid cell), which are located adjacent to the telomeres at the tops of chromosomes 2 and 4 in the ecotypes Columbia and Landsberg erecta (26, 28). A. arenosa was found to have 12 NORs among its 32 chromosomes (Fig. 1 C and D). Because many Arabidopsis species have 16 chromosomes per diploid genome (33), A. arenosa is presumed to be a tetraploid (2n = 4x = 32, where n equals the chromosome number in gametes and x represents the fundamental chromosome number). Hence, A. arenosa has three NORs per 1x complement of chromosomes, whereas A. thaliana has only two.
Fig. 1.
FISH to mitotic metaphase cells from A. thaliana ecotype No-0 (A and B) and A. arenosa strain 3651 (C and D). An A. thaliana-specific rRNA gene IGS probe (pAt2) was used in A, whereas an A. arenosa IGS probe (pCaIGS) was used in C (red FISH signals). DAPI staining shows 10 chromosomes for A. thaliana (B) and 32 for A. arenosa (D). (Bar = 5 μm.)
Fig. 2.
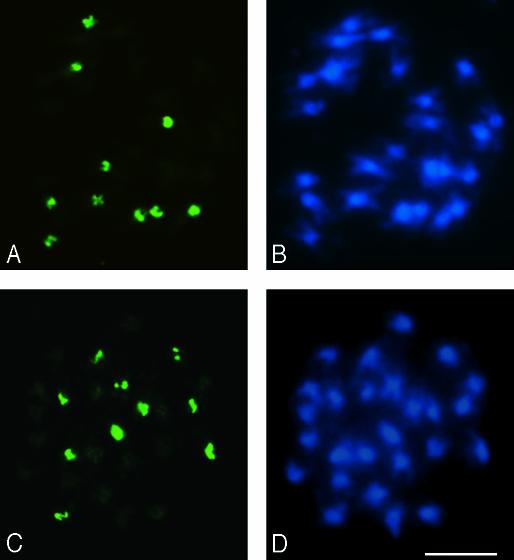
Metaphase cells of A. suecica strains LC1 (A and B) and 9502 (C and D) after in situ hybridization with an A. thaliana-specific centromere probe (pARR20-1). Ten FISH signals (green), the diploid A. thaliana (2n = 2x = 10) number, are observed in both strains (A and C). Sixteen unlabeled chromosomes visualized with DAPI (B and D) correspond to the expected 2x A. arenosa (2n = 4x = 32) chromosome number. (Bar = 5 μm.)
A. suecica has 26 chromosomes, as seen by DAPI staining in LC1 and 9502 (Fig. 2 B and D), the expected number for an allotetraploid (amphidiploid) possessing a 2x genome complement from both A. thaliana (10 chromosomes) and A. arenosa (16 chromosomes). FISH analysis using a probe corresponding to A. thaliana 180-bp centromere repeats confirms that 10 of the chromosomes in A. suecica are of A. thaliana origin (Fig. 2 A and C).
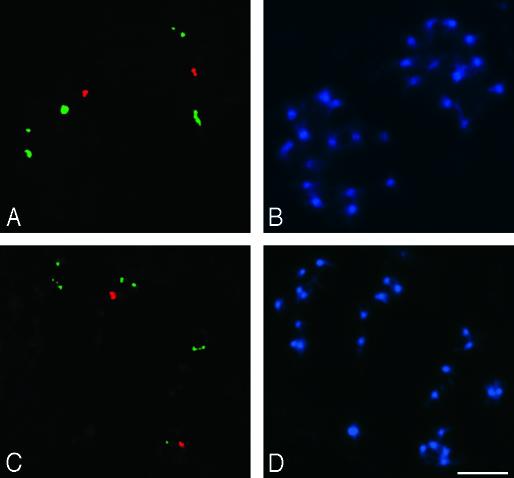
Although A. suecica strains LC1 and 9502 have the expected diploid number of A. thaliana chromosomes, they only have two A. thaliana-like NORs (Fig. 3 A and C, red FISH signals) rather than four, the expected diploid number. By contrast, six A. arenosa-derived NORs are present in both A. suecica strains (Fig. 3 A and C, green FISH signals), matching the expected number for a 2x complement of A. arenosa chromosomes. These results show that one pair of NORs of A. thaliana origin has been lost during evolution in both A. suecica strains examined. Based on linkage to cytogenetic and molecular markers, the A. thaliana NOR pair that has been retained in A. suecica is NOR4 (O.P., N.N., M.S., R.J.L., M. S. Lewis, C.S.P., and W.V., unpublished data).
Fig. 3.
Metaphase A. suecica LC1 cells (A and B) and 9502 (C and D) after in situ hybridization with rRNA gene IGS probes pAt2 (from A. thaliana, in red) and pCaIGS (from A. arenosa, in green). In both strains of A. suecica only one pair of A. thaliana NORs is present, whereas the expected six NORs from A. arenosa are retained. Chromosomes were counterstained with DAPI in B and D. (Bar = 5 μm.)
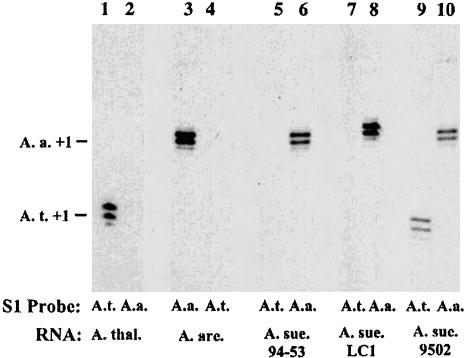
Natural Variation in Nucleolar Dominance Revealed by Transcription Analyses. Transcription of A. thaliana- and A. arenosa-derived rRNA genes in A. suecica strains LC1, 9502, and 94-53 was examined by using the S1 nuclease protection assay to determine steady-state transcript levels (Fig. 4). Total RNA isolated from each strain was split into aliquots and hybridized in separate reactions to radioactively labeled oligonucleotide probes that were specific for A. thaliana or A. arenosa rRNA transcripts initiated at the correct promoter start site (defined as +1). After digestion of the resulting RNA–DNA hybrids by S1 nuclease, the digestion products were subjected to denaturing PAGE and autoradiography. The A. thaliana probe detects A. thaliana rRNA gene transcripts but not A. arenosa transcripts (compare lanes 1 and 4 in Fig. 4). The A. arenosa probe is equally species-specific in control reactions (compare lanes 2 and 3 in Fig. 4). In A. suecica strains LC1 and 94-53, transcripts from the A. arenosa rRNA genes are abundant (lanes 6 and 8), but A. thaliana rRNA gene transcripts are undetectable by S1 nuclease protection (lanes 5 and 7). A. thaliana rRNA gene transcripts are readily detected in LC1 if plants are treated with chemical inhibitors of cytosine methylation or histone deacetylation (ref. 15; R.J.L. and C.S.P., unpublished data), indicating that the lack of A. thaliana transcript signal is due to rRNA gene silencing and not to probe–RNA transcript incompatibilities. rRNA transcripts of both A. thaliana and A. arenosa are detected in A. suecica strain 9502 (Fig. 4, lanes 9 and 10), which is true in all individuals studied over multiple generations (data not shown). These data indicate that nucleolar dominance does not occur in all A. suecica strains but exhibits natural variation as a trait.
Fig. 4.
Natural variation in nucleolar dominance in A. suecica. RNA isolated from three strains of A. suecica (A. sue.), A. thaliana (A. thal.), and A. arenosa (A. are.) was tested for A. thaliana or A. arenosa pre-rRNA transcripts by using an S1 nuclease protection assay. S1 probes were specific for A. thaliana (A.t. probe; lanes 1, 4, 5, 7, and 9) or A. arenosa (A.a. probe; lanes 2, 3, 6, 8, and 10) rRNA genes, as shown by the controls in lanes 1–4. In A. suecica strain 9502, A. arenosa and A. thaliana rRNA transcripts are equally abundant, indicating codominance.
Variation in NOR Chromatin Topology Correlates with Variation in Gene Expression. Variation in nucleolar dominance in A. suecica strains LC1 and 9502 provided a unique opportunity to compare homologous NORs in their active and inactive states, an evaluation not otherwise possible. These examinations revealed distinct differences in the NOR chromatin organization in interphase nuclei of LC1 and 9502 (Fig. 5). In LC1, A. arenosa-like NORs show partial decondensation at interphase, which is manifest in two ways. One manifestation is the appearance of FISH signal spots in excess of the number of NORs because of decondensation in central regions of the NORs flanked by regions that remain condensed (Fig. 5B; red signals). The other manifestation of decondensation is reduced FISH-staining intensity overall compared with condensed mitotic NORs (compare red signals in Fig. 5B with the green signals in Fig. 3A). By contrast to A. arenosa-derived NORs, the two A. thaliana NORs in A. suecica strain LC1 are consistently condensed into two discrete knobs (Fig. 5 A and B, green signals) that dwarf the red A. arenosa NOR signals because of the decondensation of the latter. This FISH signal differential is not apparent at mitosis when both dominant and underdominant NORs are more condensed (compare red and green signals in Figs. 5B and 3A).
Fig. 5.
Root-tip interphase cells of A. suecica strains LC1 (A–C) and 9502 (D–F) after in situ hybridization with rRNA gene IGS probes pAt2 (from A. thaliana) and pCaIGS (from A. arenosa). (A–C) A meristematic interphase nucleus from strain LC1 shows two condensed A. thaliana-derived NORs (green signals) and partially decondensed A. arenosa NORs (red signals). (D–F) A meristematic interphase nucleus from strain 9502 shows A. thaliana NORs decondensed (red signals) and decondensed A. arenosa NORs (green signals). (C and F) FISH and DAPI signals are superimposed; n denotes nucleoli. (Bar = 5 μm.)
Unlike A. suecica strain LC1, the A. thaliana-like NORs in interphase nuclei of A. suecica strain 9502 are partially decondensed, resulting in four FISH signals due to decondensation of central regions of the NORs (Fig. 5 D and E, red signals). On examining ≈120 cells in each strain (LC1 and 9502), scoring the number of A. thaliana NOR FISH signals per cell, and comparing the resulting distributions, strain 9502 shows a significant increase in NOR decondensation relative to strain LC1 (Student's t test P < 0.01; data not shown). Combined with the S1 nuclease protection data of Fig. 4, these data indicate that partial NOR decondensation is a cytogenetic manifestation of active rRNA gene transcription at an NOR.
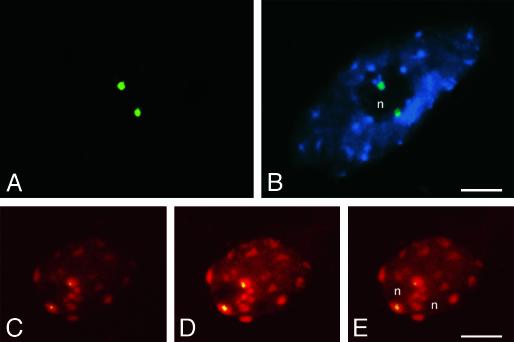
Inactive NORs Associate with the Nucleolus. After staining of interphase chromatin with DAPI or propidium iodide, nucleoli appear as dark, unstained regions (see Figs. 5 and 6). The inactive A. thaliana-derived NORs in A. suecica strain LC1 are consistently found in association with nucleoli, as is seen most clearly in Fig. 6B. As further evidence, Fig. 6 C–E shows consecutive confocal sections of a single LC1 nucleus, confirming, in a third dimension, that A. thaliana-derived NORs localize to the periphery of the nucleolus. This association of inactive A. thaliana-derived NORs with the nucleolus in LC1 occurred in 84% of meristematic root-tip cells and 76% of differentiated root-tip cells (50 of each cell type examined). Collectively, our data suggest that nucleolar association cannot be taken as evidence of transcriptional activity of an NOR, in agreement with a careful study of NORs in cultured mammalian cells (22).
Fig. 6.
Both active and inactive NORs associate with the nucleolus. (A and B) A differentiated root-tip cell from strain LC1 showing the two A. thaliana-derived NORs (green signals; pAt2 FISH probe) associated with the nucleolus (n), which appears as a dark region unstained by DAPI (FISH and DAPI signals are superimposed in B). (C–E) Three optical sections obtained by confocal microscopy of a meristematic root-tip cell of strain LC1, further demonstrating the association of inactive A. thaliana-derived NORs with nucleoli. A. thaliana-derived NORs appear yellow because of the superimposition of green FISH signals (pAt2 probe) and red, propidium iodide-stained chromatin. Note that heterochromatic regions provide the brightest red signals. NOR signals in C–E appear smaller than in A and B because they are individual confocal sections; integrated images of many sections would be larger. (Bar = 5 μm.)
Genetic Similarity of A. suecica Strains Displaying Variation in Nucleolar Dominance. Our detection of natural variation in nucleolar dominance prompted us to evaluate the degree of genetic similarity in A. suecica strains 9502, LC1, and 94-53 by using AFLP analysis of genomic DNA (Fig. 7). By using five different primer pair combinations, AFLP indicated that all three strains of A. suecica are genetically very similar, with only ≈4% (14 of 327) of all amplified bands revealing a polymorphism in any pairwise combination. These data suggest that variation in nucleolar dominance is likely to result from relatively subtle genetic or epigenetic variation.
Fig. 7.
AFLP analysis of A. suecica strains showing natural variation in nucleolar dominance. Genomic DNA fragments of A. suecica strains LC1, 94-53, and 9502 were analyzed by using five different selective primers (M-CAA, M-CAC, M-CAT, M-CTA, and M-CTT). Bands of distinct mobility that are not common to all three strains are marked by arrows. Bands that differ only in intensity are not marked because these might result only from differences in restriction endonuclease digestion efficiency, possibly due to cytosine methylation, an epigenetic modification.
Discussion
A previous study showed that nucleolar dominance occurs both in a natural strain of A. suecica and in synthetic A. suecica-like allotetraploids created in the laboratory (15). In the current study, we show that nucleolar dominance is not a fundamental trait of A. suecica as a species. We have exploited this natural variation in nucleolar dominance to deduce the relationships among NOR chromatin topology, NOR–nucleolus association, and rRNA gene transcription. Our observations suggest that transcriptionally inactive NORs are highly condensed both at interphase and at metaphase and thus appear as bright spots after FISH. By contrast, active NORs appear to be composed of condensed knobs that are interconnected by decondensed regions of rRNA gene chromatin. One interpretation of these data is that only a subset of the rRNA genes is transcribed even at dominant NORs, and this subset is the fraction that makes up the decondensed NOR chromatin. Indeed, in wheat–rye chromosome addition lines, dominant wheat NORs also display condensed knobs interspersed with decondensed rRNA gene chromatin (A. P. Santos, M.S., N.N., and W.V., unpublished data). In these addition lines, only the decondensed regions of wheat NORs incorporate BrUTP, indicating that these intervals contain transcriptionally active rRNA genes. Likewise, investigations of diploid rye have shown that condensed rRNA gene chromatin is located at the periphery of the nucleolus, whereas decondensed chromatin extends into the central portions of the nucleolus where rRNA transcription takes place (34). Condensed regions are thus thought to be portions of the NOR in which transcriptionally inactive rRNA genes are packaged into heterochromatin.
Although it is difficult to quantify the number of rRNA genes located within the condensed and decondensed portions of the NOR, visual inspection of FISH images suggests that only a small fraction of the A. thaliana rRNA genes are decondensed, and are thus presumed to be transcriptionally active, in A. suecica strain 9502. This finding may not be surprising given the large number of rRNA genes in plants compared with budding yeast, Drosophila, Xenopus, or mammals, whose haploid rRNA gene numbers range between 150 and 400 genes. In yeast and mouse cells, psoralen-crosslinking experiments suggest that only one-third to one-half of the rRNA genes are transcribed (which makes them accessible to psoralen) in actively growing cultured cells (refs. 35–38; reviewed in ref. 39). A. thaliana has relatively few rRNA genes compared with other plants, yet still has an estimated 750 rRNA genes per haploid genome. If only 75–200 rRNA genes need to be transcriptionally active (extrapolating from yeast and mouse, respectively), one might expect only ≈10–27% of the rRNA genes in a diploid A. thaliana nucleus to be active. In an allotetraploid hybrid such as A. suecica strain 9502, which transcribes rRNA genes of both parental species, the fraction of the A. thaliana-derived rRNA genes that are transcribed may be lower still. This finding may explain why the condensed knobs are the most prevalent feature of A. thaliana-derived NORs in A. suecica strain 9502, despite the transcription of A. thaliana rRNA genes in this strain.
As shown in Fig. 6, the A. thaliana-derived NORs associate with the nucleolus in A. suecica strain LC1 even though no transcription from these NORs can be detected by using the S1 nuclease protection assay. An interesting parallel in mammals is that transcriptionally inactive human NORs in mouse–human cell hybrids colocalize to the nucleolus with transcriptionally active mouse NORs (22). It is possible that trace levels of transcription play some role in NOR–nucleolus associations. Alternatively, positional cues other than rRNA gene transcription may be key. One possibility is that sequence-specific DNA-binding proteins, rather than rRNA transcription, provide signals for nucleolus assembly or NOR–nucleolus association.
The basis for natural variation in nucleolar dominance is not clear. Although A. suecica strains LC1 and 9502 differ in terms of nucleolar dominance, they are genetically similar. One large-scale similarity is that one pair of A. thaliana-derived NORs has been lost in both LC1 and 9502, the missing NORs being NOR2 in both cases (O.P., N.N., M.S., R.J.L., M. S. Lewis, C.S.P., and W.V., unpublished data). Likewise, the A. thaliana-derived NORs in both strains appear to be similar in size based on metaphase FISH signal intensity, are similarly positioned near the ends of their respective chromosomes, and are similarly associated with nucleoli regardless of transcriptional activity. On a finer scale, AFLP revealed that only ≈4% of amplified bands showed any difference in size in the two strains. For comparison, AFLP analysis of 38 A. thaliana ecotypes revealed that, on average, ≈17% of AFLP bands are polymorphic between ecotypes, with the most similar pair of ecotypes revealing polymorphic bands at a frequency of ≈4% (40). Hence, the A. suecica strains we have examined are genetically very similar, comparable with the similarity of the most closely related ecotypes of A. thaliana. Collectively, these observations suggest that natural variation in nucleolar dominance in A. suecica is likely to result from subtle genetic (or epigenetic) variation rather than dramatic genome restructuring events. This finding suggests that it might be possible to identify different ecotypes of the progenitor species that program different nucleolar dominance outcomes when crossed to re-create the hybrid. If such variation can be found among A. thaliana parents, in particular, the genetic tools available could facilitate identification of genes controlling nucleolar dominance.
One clue concerning the types of regulatory genes that might explain natural variation in nucleolar dominance is the fact that decondensation of A. thaliana-derived NORs in A. suecica strain 9502 (Fig. 5) is also accompanied by a greater degree of decondensation among the A. arenosa-derived NORs when compared with strain LC1. Whereas the average number of condensed FISH signals observed for the dominant A. arenosa-like NORs in 136 interphase cells of strain LC1 was 5.9 ± 1.7 (mean ± SE), the average number observed in 120 interphase cells of strain 9502 was 8.7 ± 2.0, a significant difference (P < 0.01; Student's t test). Examination of chromatin visualized by DAPI, which stains the condensed heterochromatin most brightly, failed to reveal obvious differences in overall chromatin condensation between the two strains. Therefore, one possibility is that strain 9502 has decreased activity for one or more proteins that affect NOR or rRNA gene condensation without acting throughout the whole genome. Future studies are needed to elucidate whether such changes in NOR chromatin topology are a cause or a consequence of changes in rRNA gene transcription.
Acknowledgments
We thank Steve O'Kane (University of Northern Iowa, Cedar Falls) and Luca Comai (University of Washington, Seattle) for A. suecica and A. arenosa seeds, and Eric Richards (Washington University) for the A. thaliana centromere clone. Augusta Barão provided excellent cytogenetics assistance as did Margarida Delgado (Viegas laboratory) and Rui Malhó (Faculdade de Ciências, Universidade de Lisboa, Lisbon) for confocal analyses. This work was supported by National Institutes of Health Grant R01-GM60380 (to C.S.P.) and by Fundação para a Ciência e Tecnologia Project POCTI/BCI/38557/2001 (to N.N.). Publication costs were defrayed, in part, by a research prize to R.J.L. from the Jean Lowenhaupt Botany Fund.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AFLP, amplified fragment-length polymorphism; FISH, fluorescence in situ hybridization; NOR, nucleolus organizer region; DAPI, 4′,6′-diamidino-2-phenylindole hydrochloride; IGS, intergenic spacer.
References
- 1.Reeder, R. H. (1985) J. Cell Biol. 101, 2013–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pikaard, C. S. (2000) Plant Mol. Biol. 43, 163–177. [DOI] [PubMed] [Google Scholar]
- 3.Neves, N., Castilho, A., Silva, M., Heslop-Harrison, J. S. & Viegas, W. (1997) in Chromosomes Today, ed. Puertas, M. J. (Chapman & Hall, London), Vol. 12, pp. 182–200. [Google Scholar]
- 4.Viegas, W., Neves, N., Caperta, A., Silva, M. & Morais-Cecílio, L. (2002) Curr. Genomics 3, 563–576. [Google Scholar]
- 5.Navashin, M. (1934) Cytologia 5, 169–203. [Google Scholar]
- 6.McClintock, B. (1934) Z. Zellforsch. Mikrosk. Anat. 21, 294–328. [Google Scholar]
- 7.Heitz, E. (1931) Planta 15, 495–505. [Google Scholar]
- 8.Ritossa, F. M. & Spiegelman, S. (1965) Proc. Natl. Acad. Sci. USA 53, 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace, H. & Birnstiel, M. L. (1966) Biochim. Biophys. Acta 114, 296–310. [DOI] [PubMed] [Google Scholar]
- 10.Phillips, R. L., Kleese, R. A. & Wang, S. S. (1971) Chromosoma 36, 79–88. [Google Scholar]
- 11.Honjo, T. & Reeder, R. H. (1973) J. Mol. Biol. 80, 217–228. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Z. J. & Pikaard, C. S. (1997) Genes Dev. 11, 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace, H. & Langridge, W. H. R. (1971) Heredity 27, 1–13. [Google Scholar]
- 14.Pikaard, C. S. (2000) Trends Genet. 16, 495–500. [DOI] [PubMed] [Google Scholar]
- 15.Chen, Z. J., Comai, L. & Pikaard, C. S. (1998) Proc. Natl. Acad. Sci. USA 95, 14891–14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comai, L., Tyagi, A. P., Winter, K., Holmes-Davis, R., Reynolds, S. H., Stevens, Y. & Byers, B. (2000) Plant Cell 12, 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanfstingl, U., Berry, A., Kellogg, E. A., Costa, J. T., III, Rudiger, W. & Ausubel, F. M. (1994) Genetics 138, 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Kane, S., Schaal, B. & Al-Shehbaz, I. (1995) Syst. Bot. 21, 559–566. [Google Scholar]
- 19.Jones, G. H. & Heslop-Harrison, J. S. (2000) in Arabidopsis, a Practical Approach, ed. Wilson, Z. A. (Oxford Univ. Press, Oxford), pp. 105–124.
- 20.Vongs, A., Kakutani, T., Martienssen, R. A. & Richards, E. J. (1993) Science 260, 1926–1928. [DOI] [PubMed] [Google Scholar]
- 21.Doelling, J. H., Gaudino, R. J. & Pikaard, C. S. (1993) Proc. Natl. Acad. Sci. USA 90, 7528–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sullivan, G. J., Bridger, J. M., Cuthbert, A. P., Newbold, R. F., Bickmore, W. A. & McStay, B. (2001) EMBO J. 20, 2867–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maluszynska, J. & Heslop-Harrison, J. S. (1991) Plant J. 1, 159–166. [Google Scholar]
- 24.Albini, S. M. (1994) Plant J. 5, 665–672. [Google Scholar]
- 25.Sears, L. M. S. & Lee-Chen, S. (1970) Can. J. Genet. Cytol. 12, 217–223. [Google Scholar]
- 26.Copenhaver, G. P., Doelling, J. H., Gens, J. S. & Pikaard, C. S. (1995) Plant J. 7, 273–286. [DOI] [PubMed] [Google Scholar]
- 27.Copenhaver, G. P. & Pikaard, C. S. (1996) Plant J. 9, 273–282. [DOI] [PubMed] [Google Scholar]
- 28.Copenhaver, G. P. & Pikaard, C. S. (1996) Plant J. 9, 259–272. [DOI] [PubMed] [Google Scholar]
- 29.Ambros, P. & Schweizer, D. (1976) Arabidopsis Info. Serv. 13, 167–171. [Google Scholar]
- 30.Bauwens, S., Van Oostveldt, P., Engler, G. & Van Montague, M. (1991) Chromosoma 101, 41–48. [DOI] [PubMed] [Google Scholar]
- 31.Koornneef, M., van Eden, J., Hanhart, C. J., Braaksma, F. J. & Feenstra, W. J. (1983) J. Hered. 74, 265–272. [Google Scholar]
- 32.Henikoff, S. & Comai, L. (1998) Genetics 149, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Shehbaz, I. A. & O'Kane, S. L. (2002) in The Arabidopsis Book, ed. Meyerowitz, E. M. (Am. Soc. Plant Biologists, Rockville, MD), 10.1199/tab. 0009. Available at www.aspb.org/publications/arabidopsis. Accessed August 29, 2003.
- 34.Caperta, A. D., Neves, N., Morais-Cecilio, L., Malho, R. & Viegas, W. (2002) J. Cell Sci. 115, 2839–2846. [DOI] [PubMed] [Google Scholar]
- 35.Conconi, A., Widmer, R. M., Koller, T. & Sogo, J. M. (1989) Cell 57, 753–761. [DOI] [PubMed] [Google Scholar]
- 36.Dammann, R., Lucchini, R., Koller, T. & Sogo, J. M. (1993) Nucleic Acids Res. 21, 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.French, S. L., Osheim, Y. N., Cioci, F., Nomura, M. & Beyer, A. L. (2003) Mol. Cell. Biol. 23, 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandmeier, J. J., French, S., Osheim, Y., Cheung, W. L., Gallo, C. M., Beyer, A. L. & Smith, J. S. (2002) EMBO J. 21, 4959–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grummt, I. & Pikaard, C. S. (2003) Nat. Rev. Mol. Cell Biol. 4, 641–649. [DOI] [PubMed] [Google Scholar]
- 40.Miyashita, N. T., Kawabe, A. & Innan, H. (1999) Genetics 152, 1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]