Abstract
Microtubules mediate mitochondrial distribution in the yeast Schizosaccharomyces pombe and many higher eukaryotic cells. In higher eukaryotes, kinesin motor proteins have been shown to transport mitochondria along microtubules, but the nature of the mitochondria–microtubule interactions in S. pombe has not been explored. By time lapse, total internal reflection fluorescence microscopy, or spinning-disk confocal microscopy, mitochondria appeared to be both tethered to ends and bound laterally along the sides of microtubules. Mitochondrial tubules extended and retracted when attached to the tips of elongating or shortening microtubules, respectively, but translocation along established microtubules was never observed. Mitochondria that were not associated with microtubules were largely immobile until they were “captured” by a growing microtubule. In mitotic cells, a portion of the mitochondria was tethered to the spindle-pole bodies and moved to the cellular ends during spindle elongation. This association may be important for organelle inheritance during cell division. Thus, in contrast to kinesin-mediated transport used by higher eukaryotes, mitochondrial motility and distribution in fission yeast are driven largely by microtubule polymerization and the elongation of the mitotic spindle.
The distribution of mitochondria throughout the cytoplasm in most eukaryotic cells depends on the cytoskeleton (1). In higher eukaryotes, the microtubule-based motor protein kinesin as well as kinesin-related proteins (e.g., KIF1B) play a central role in this process by transporting mitochondria along cytoplasmic microtubules (2, 3). Microtubules also mediate the distribution of mitochondria in the fission yeast Schizosaccharomyces pombe (4). Mitochondria colocalize with microtubules during interphase of the fission yeast cell cycle (4, 5), and certain mutations in tubulin genes cause dramatic changes in mitochondrial distribution in mutant S. pombe cells, with mitochondria becoming aggregated and asymmetrically positioned (4). However, mitochondrial distribution in fission yeast does not appear to depend on the activity of klp3, the major cytoplasmic kinesin (6), or the function of other kinesin-like proteins (B. A. Weir and M.P.Y., unpublished observations). Thus, the mechanism by which microtubules determine mitochondrial positioning has remained obscure.
To examine the dynamic relationships of mitochondria and microtubules in S. pombe cells, we developed a strain of fission yeast with distinct fluorescent proteins labeling mitochondria and microtubules and imaged their behavior by using total internal-reflection fluorescence microscopy on interphase cells and spinning-disk confocal fluorescence microscopy on mitotic cells. This analysis revealed that mitochondria do not translocate along microtubules as they do in higher eukaryotic cells. Rather, mitochondria bind to microtubules, and the polymerization and depolymerization of microtubules drive an extension and retraction, respectively, of mitochondrial tubules during interphase. Additionally, association with the mitotic spindle poles facilitates mitochondrial distribution during mitosis.
Materials and Methods
Yeast Strains and Sample Preparation. A S. pombe strain (#641; ars1::nmt-atb1-GFP:LEU2, ade6-m210, his3-D1, leu1–32, ura4-D18, h–) expressing a GFP-α1-tubulin fusion protein was the gift of D. Williams and D. McIntosh (University of Colorado, Boulder). An S. pombe strain expressing Discosoma red fluorescent protein (RFP) targeted to mitochondria was created as follows. DNA sequences corresponding to the mitochondrial targeting signal domain of Saccharomyces cerevisiae cytochrome oxidase subunit IV (pCOX4) were generated by PCR by using primers SS-MfeI-f (CAATTGATGCTTTCACTACGTCAATC) and SS-PacI-r (TTAATTAAAGTTTTCACCACGGGTTTTTGC) and S. cerevisiae genomic DNA as template. Sequences corresponding to RFP were generated by PCR by using primers RFP-PacI-f (TTAATTAAGACCATGATTACGCCAAGC) and RFP-N/M-r (GCGGCCGCCAATTGCTAAAGGAACAGATGGTGGC) and plasmid pDsRed (CLONTECH) as template. The pCOX4 and RFP PCR products were cloned into the TOPO vector pCR2.1 (Invitrogen) to generate pCR-C4 and pCR-RFP, respectively. The PacI–NotI fragment from pCR-RFP (containing RFP) was subcloned into the PacI and NotI sites of pCR-C4 to yield pCR-C4RFP. The PsaI–SacI fragment containing the S. pombe nmt promoter was isolated from plasmid pREP3X (7) and cloned into the PsaI and SacI sites of plasmid pJK148 (8) to yield pJK148-nmt. The MfeI fragment containing the pCOX4-RFP gene fusion sequences was isolated from plasmid pCR-C4RFP, the ends were filled with Klenow, and the fragment was ligated into the SmaI site of pJK148-nmt. The resulting plasmid was cut with NruI and transformed into S. pombe strain FY261 (ade6-m216, ura4-D18, leu1–32, can1–1, h+; obtained from S. Forsburg, Salk Institute, La Jolla, CA). A stable leu+ transformant containing the nmt-pCOX4-RFP integrated at the leu1 locus was designated strain MYP100 (nmt-pCOX4-RFP:leu1+, ade6-m216, ura4-D18, leu1–32, can1–1, h+). MYP101, a strain harboring both GFP-α1-tubulin and pCOX4-RFP was derived as a haploid segregrant from a cross of strain 641 with strain MYP100.
Cells to be analyzed microscopically were cultured first for 24–36 h on liquid Edinburgh minimal medium (EMM) (9) without thiamine at 25°C and then for an additional 16–24 h at 25°C after addition of thiamine to 5 μg/ml. Cells were immobilized on fused silica slides with 1% low melting agarose in EMM, and, for time-lapse total internal-reflection fluorescence (TIRF) microscopy, coverslips were sealed with rubber cement.
Microscopic Analysis. The total internal reflection fluorescence microscope used in these studies has been described (10) but was modified for simultaneous viewing of GFP and RFP. Both fluorescent proteins were excited with the 488-nm line of an argon laser. The GFP and RFP emission was then split by using a dichroic mirror, and the RFP emission was translated by using mirrors before it was recombined with the GFP emission. This allowed offset GFP and RFP images to be projected simultaneously to a cooled charge-coupled device camera (Sensicam, Cooke Corp., Tonawanda, NY). This arrangement is similar to the design by Kinosita (11) for projecting the donor and acceptor fluorescence simultaneously to a camera in a microscope designed for fluorescence resonance energy transfer. Images were captured by using imaging workbench software (Axon Instruments, Foster City, CA) with images acquired for 3 to 5 s at intervals of 15 or 20 s.
Confocal microscopy was performed by using a Yokogawa spinning-disk confocal scanhead (Solamere Technology Group, Salt Lake City, UT) attached to an inverted fluorescence microscope (Axiovert 200 m, Zeiss). Both GFP and RFP were excited with the 488-nm line of an argon laser. GFP and RFP were imaged sequentially, by using specific emission filters (GFP: HQ530/60, RFP: HQ605/75, Chroma Technology, Brattleboro, VT) mounted in a filter wheel. Images were acquired by using a Mega 10 intensified charge-coupled device camera (Stanford Photonics, Palo Alto, CA). The software package qed (QED Imaging, Pittsburgh) was used to control the camera, laser shutter, microscope, and filter wheel.
Results
To examine the behavior of mitochondria and microtubules in vivo, S. pombe cells expressing GFP-tubulin and RFP targeted to mitochondria via a fusion with precytochrome oxidase subunit IV were initially observed by TIRF microscopy. The GFP and RFP images were offset from one another before projection onto a cooled charge-coupled device camera to allow simultaneous observation of both fluorophores (see Materials and Methods). Total internal reflection microscopy provides very low background, but fluorescence excitation is restricted to a few hundred nanometers from the surface. Nevertheless, we could observe images of microtubules and mitochondria in most interphase cells. This microscopic method was primarily advantageous in allowing imaging of mitochondria and microtubules for extended time periods (even with continuous illumination) with no apparent perturbation of cellular function, as evidenced by continued dynamic behavior of microtubules. In contrast, we found that even a few minutes of continuous light exposure by wide-field or conventional confocal epifluorescence illumination resulted in toxicity and perturbations of microtubule dynamics.
One indication of cellular vitality during sample analysis was the robust growth and depolymerizaton of microtubules that displayed an average polymerization rate of 1.97 ± 0.58 μm/min (n = 40). This rate is very similar to that described for S. pombe cells in previous studies using a different imaging methodology (12, 13).
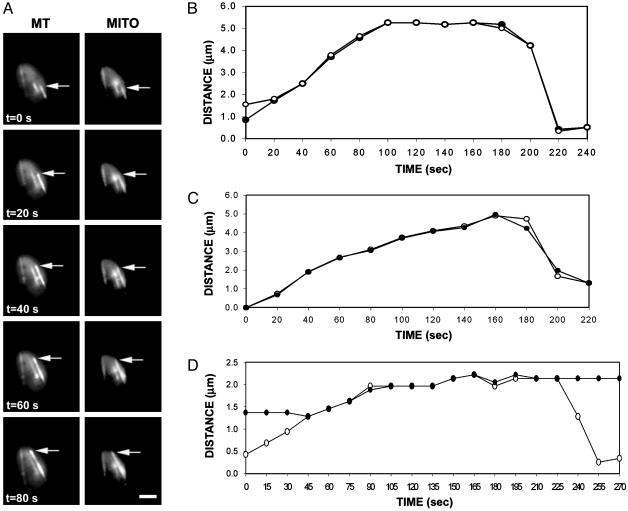
Time-lapse image sequences of cells revealed movement of mitochondrial “tubules” concomitant with microtubule polymerization (Fig. 1 and Movie 1, which is published as supporting information on the PNAS web site, www.pnas.org). These mitochondrial tubules appeared to be associated with growing ends of microtubules and to “elongate” as microtubule polymerization progressed (Fig. 1 A). The parallel dynamic behavior of mitochondrial tubules and microtubules is illustrated in the life-history plots in Fig. 1 B–D. In addition to attachment and movement along microtubule tips, mitochondria also were frequently aligned along the length of microtubules and appeared to be bound laterally. The distinct identities of microtubules and mitochondria detected with TIRF microscopy were confirmed by using cells expressing only a single fluorescent protein (data not shown) and by the distinctive and unique mitochondrial or microtubule profiles observed in some cellular images (Figs. 2 and 3).
Fig. 1.
Mitochondria move concomitantly with microtubule polymerization. Cells of S. pombe strain MYP101 induced for expression of GFP-tubulin and RFP targeted to mitochondria were imaged for GFP fluorescence (microtubules) and RFP fluorescence (mitochondria) using TIRF microscopy. (A) Microtubules (MT) and mitochondria (MITO) are shown in successive time-lapse images of a portion of a single cell imaged for 5 s at 20-s intervals. Arrows indicate the growing end of the microtubule and the end of the corresponding mitochondrial tubule. (Bar = 2 μm.) (B–D) Coordinate dynamics of microtubules and mitochondria. The distance of the microtubule tip (open symbols) and the tip of an associated mitochondrial tubule (filled symbols) (relative to an arbitrary position of near the tip of the mitochondrial tubule or the microtubule at time 0) plotted against time. Shown are plots for three distinct microtubule–mitochondrial interactions. In D, the mitochondria are not associated with the microtubule at the beginning and end of the time course.
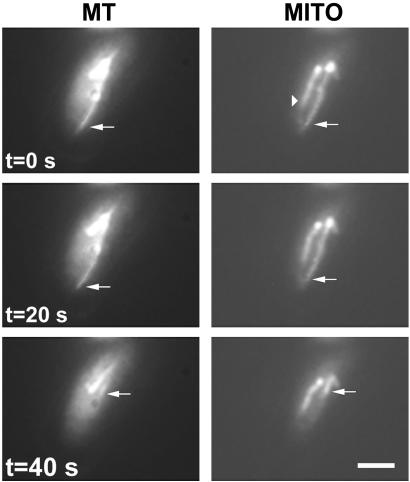
Fig. 2.
Mitochondrial repositioning mediated by microtubule depolymerization. A cell of strain MYP101 was imaged for microtubules (MT) and mitochondria (MITO) as described for Fig. 1. A microtubule undergoing catastrophe and an associated mitochondrion are indicated by the arrows. Also labeled (arrowhead) is a mitochondrial tubule not associated with a microtubule. (Bar = 2 μm.)
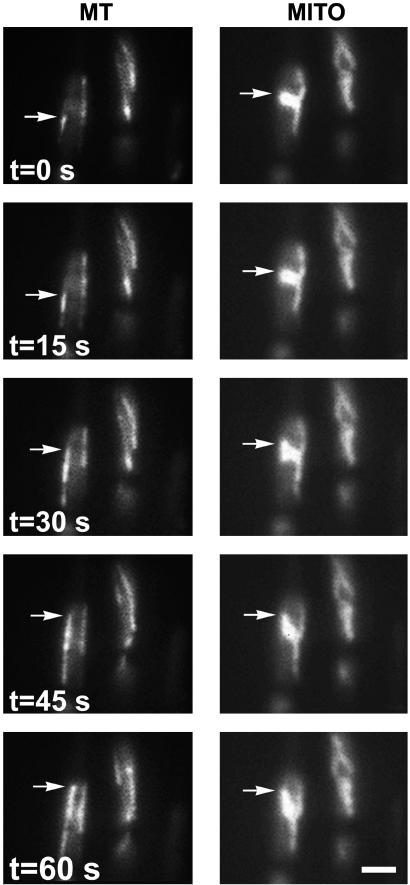
Fig. 3.
Capture of a mitochondrion by a growing microtubule. Time-lapse images of microtubules (MT) and mitochondria (MITO) were collected as described for Fig. 1, except that they were acquired at 15-s intervals. Arrows indicate a polymerizing microtubule and a portion of a mitochondrial tubule that is repositioned after becoming associated with the microtubule. (Bar = 2 μm.)
Mitochondrial movement also accompanied microtubule depolymerization. Some mitochondrial tubules appeared to retract or rapidly reposition with the depolymerization of an associated microtubule (Fig. 2). The similar rates of microtubule depolymerization and mitochondrial retraction (Fig. 1 B–D) suggested that the mitochondria could remain bound to the end of a depolymerizing microtubule. However, in other instances, a mitochondrion appeared to lose its association with a microtubule as depolymerization occurred, with the mitochondrion remaining in a fixed position in the cell (see Fig. 1D).
Mitochondria not associated with microtubules displayed little progressive linear movement (Fig. 2). Additionally, the progressive extension or retraction of mitochondrial tubules was not apparent in cells treated with the microtubule inhibitor thiabendazole (data not shown), although mitochondria tended to fragment during treatment with this drug. Importantly, the translocation of mitochondria along established or immobile microtubules was not observed in time-lapse analyses of 160 different cells.
Microtubules were occasionally observed to “capture” mitochondria (Fig. 3), leading to the movement or repositioning of mitochondrial tubules that had remained stationary before microtubule association (Fig. 3). Such capture appeared to involve an interaction between the growing end of a polymerizing microtubule and the mitochondrial surface.
Mitochondrial behavior in mitotic S. pombe cells was analyzed by spinning-disk confocal microscopy, because TIRF microscopy failed to detect mitotic spindles that lie too far from the cell surface to be excited by the evanescent wave. This analysis revealed that the spindle apparatus facilitates mitochondrial distribution during mitosis (Fig. 4 and Movie 2, which is published as supporting information on the PNAS web site). After entering mitosis, mitochondria became stably tethered to each of the spindle poles in all cells examined (n > 100). Transient interactions with the highly dynamic astral microtubules were also observed. As mitosis proceeded and the spindle elongated, mitochondria were moved to opposite ends of the cell via their attachment to the spindle pole bodies (Fig. 4 and Movie 2). In occasional cells, mitosis became stalled at an early stage with short mitotic spindles (possibly due to illumination-induced photodamage), yet some mitochondria remained closely associated with the spindle poles for extended periods. The concomitant Brownian motion of these spindle poles and associated mitochondria provided further evidence of a persistent attachment of mitochondria to the spindle pole (not shown). Additionally, a subset of the cell's mitochondria frequently appeared immobile and unassociated with the spindle apparatus and remained near the center of the cell (93 of 100 cells examined had one or a few mitochondria remaining near the cell center at the time of cytokinesis). These may represent mitochondria that did not have an opportunity to form interactions with the spindle poles or that formed other interactions near the cell center.
Fig. 4.
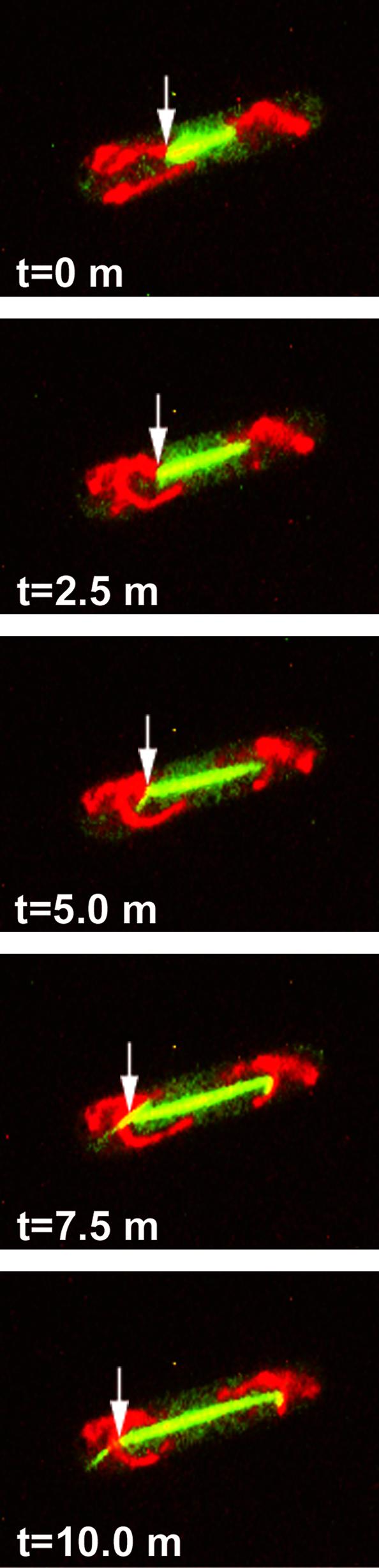
Spindle elongation transports mitochondria toward the cell tips. Cells of S. pombe strain MYP101 induced for expression of GFP-tubulin and RFP targeted to mitochondria were imaged for GFP fluorescence (green; microtubules) and RFP fluorescence (red; mitochondria) by using spinning-disk confocal microscopy. Shown are successive time-lapse images of a mitotic cell captured for 0.5 s at 2.5-min intervals. Arrows indicate one of the cell's spindle pole bodies and its association with mitochondria. Also evident (at times 5.0, 7.5, and 10.0 min) are aster microtubules projecting from spindle-pole bodies, with mitochondria apparently splayed along these transient structures.
Discussion
Our results indicate that the polymerization and depolymerization of microtubules play key roles in the positioning of mitochondria in fission yeast during interphase. This mechanism of organellar distribution exploits the active microtubule dynamics in S. pombe cells in which microtubules grow out toward the cell tips from microtubule organizing centers on or near the nuclear envelope and shrink back toward the nucleus after catastrophe (12, 13). These dynamics result in the creation of mitochondrial tubules and the extension of these tubules toward the cell tips during polymerization. This transport may supply mitochondrial metabolic functions in areas of particular cellular need or serve an important function in supplying mitochondria to the new cellular ends generated through cell division (14). Consistent with such a latter role, mitochondria become asymmetrically distributed in certain S. pombe mutants with defective microtubule function (4).
The mechanism for mitochondrial movement in fission yeast revealed by our analyses differs from the predominant mechanism of organelle movement in animal cells involving transport along microtubules by the motor proteins kinesin and cytoplasmic dynein (2, 15). Evidence of a role for these molecular motors in mitochondrial movement in S. pombe is lacking (refs. 6 and 16; B. Weir and M.P.Y., unpublished observations). However, animal cells also can use microtubule polymerization dynamics for the positioning of subcellular organelles. In particular, attachment to the growing ends of microtubules in migrating newt lung epithelial cells was found to contribute (together with molecular motor-based transport) to the movement and positioning of tubules of the endoplasmic reticulum near the cell periphery (17). A similar mechanism involving microtubule dynamics was observed for the movement of phagosomes in normal rat kidney cells in culture (18). Future studies may reveal additional cellular transport processes that depend on the polymerization and depolymerization of microtubules.
The role of the mitotic spindle apparatus in localizing mitochondria to the two cellular poles represents a previously undescribed function for this key cellular structure. Previous studies have documented an association of mitochondria and other organelles with spindle aster microtubules in other species (19, 20), but the functional significance of these associations has been unknown. Our results indicate that such an association provides one mechanism for the faithful segregation of mitochondria to opposite ends of the cell, ensuring inheritance of mitochondria by daughter cells. Further studies may reveal use of this mechanism by other cell types.
The identities of proteins that mediate attachment of mitochondria to microtubules and to the mitotic spindle poles in fission yeast cells are unknown. Our results suggest that such proteins should include species that bind to the dynamic (plus) microtubule ends, proteins that interact with the sides of interphase microtubules, and certain proteins associated with the spindle pole bodies in mitotic cells. These components could interact either directly with proteins of the mitochondrial outer membrane or through linker proteins. Previous studies have identified several proteins in S. pombe that interact with microtubules and concentrate at the cell tips [tip1p (21), tea1p (22), and tea2p (23)], but these species do not appear to play a direct role in facilitating microtubule interaction with mitochondria, because cells mutant for these components still distribute mitochondria toward the cell tips (B. Weir and M.P.Y., unpublished observations). Future genetic and biochemical approaches should lead to the identification of proteins that enable cells to couple microtubule and spindle dynamics with the productive and coordinated distribution of mitochondria.
Supplementary Material
Acknowledgments
We thank DeWight Williams and Dick McIntosh for the gift of the S. pombe strain expressing GFP-tubulin; Peter Fekkes, Wen-Yuan Hu, and Barbara Weir for assistance in development of S. pombe strains expressing RFP targeted to mitochondria; and Kurt Thorn for instrument modifications and valuable assistance with TIRF microscopy. This work was supported by grants from the National Institutes of Health (GM44614 to M.P.Y. and GM38499 to R.D.V.).
Abbreviations: RFP, Discosoma red fluorescent protein; TIRF, total internal-reflection fluorescence.
References
- 1.Yaffe, M. P. (1999) Science 283, 1493–1497. [DOI] [PubMed] [Google Scholar]
- 2.Vale, R. D. (1987) Annu. Rev. Cell Biol. 3, 347–378. [DOI] [PubMed] [Google Scholar]
- 3.Nangaku, M., Sato-Yoshitake, R., Okada, Y., Noda, Y., Takemura, R., Yamazaki, H. & Hirokawa, N. (1994) Cell 79, 1209–1220. [DOI] [PubMed] [Google Scholar]
- 4.Yaffe, M. P., Harata, D., Verde, F., Eddison, M., Toda, T. & Nurse, P. (1996) Proc. Natl. Acad. Sci. USA 93, 11664–11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanbe, T., Kobayashi, I. & Tanaka, K. (1989) J. Cell Sci. 94, 647–656. [DOI] [PubMed] [Google Scholar]
- 6.Brazer, S. C., Williams, H. P., Chappell, T. G. & Cande, W. Z. (2000) Yeast 16, 149–166. [DOI] [PubMed] [Google Scholar]
- 7.Forsburg, S. L. (1993) Nucleic Acids Res. 21, 2955–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keeney, J. B. & Boeke, J. D. (1994) Genetics 136, 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreno, S., Klar, A. & Nurse, P. (1991) Methods Enzymol. 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 10.Pierce, D. W. & Vale, R. D. (1999) Methods Cell Biol. 58, 49–73. [DOI] [PubMed] [Google Scholar]
- 11.Kinosita, K., Jr., Itoh, H., Ishiwata, S., Hirano, K., Nishizaka, T. & Hayakawa, T. (1991) J. Cell Biol. 115, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drummond, D. R. & Cross, R. A. (2000) Curr. Biol. 10, 766–775. [DOI] [PubMed] [Google Scholar]
- 13.Tran, P. T., Marsh, L., Doye, V., Inoue, S. & Chang, F. (2001) J. Cell Biol. 153, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nurse, P. (1994) Mol. Biol. Cell 5, 613–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers, S. L. & Gelfand, V. I. (2000) Curr. Opin. Cell Biol. 12, 57–62. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto, A., West, R. R., McIntosh, J. R. & Hiraoka, Y. (1999) J. Cell Biol. 145, 1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waterman-Storer, C. M. & Salmon, E. D. (1998) Curr. Biol. 8, 798–806. [DOI] [PubMed] [Google Scholar]
- 18.Blocker, A., Griffiths, G., Olivo, J. C., Hyman, A. A. & Severin, F. F. (1998) J. Cell Sci. 111, 303–312. [DOI] [PubMed] [Google Scholar]
- 19.Aist, J. R. & Bayles, C. J. (1991) Eur. J. Cell Biol. 56, 358–363. [PubMed] [Google Scholar]
- 20.Pereira, A. J., Dalby, B., Stewart, R. J., Doxsey, S. J. & Goldstein, L. S. (1997) J. Cell Biol. 136, 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunner, D. & Nurse, P. (2000) Cell 102, 695–704. [DOI] [PubMed] [Google Scholar]
- 22.Behrens, R. & Nurse, P. (2002) J. Cell Biol. 157, 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Browning, H., Hayles, J., Mata, J., Aveline, L., Nurse, P. & McIntosh, J. R. (2000) J. Cell Biol. 151, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.