Abstract
Cytoplasmic mRNA localization provides a means of generating cell asymmetry and segregating protein activity. Previous studies have identified two mRNAs that localize to the bud tips of the yeast Saccharomyces cerevisiae. To identify additional localized mRNAs, we immunoprecipitated the RNA transport components She2p, She3p, and Myo4p and performed DNA microarray analysis of their associated RNAs. A secondary screen, using a GFP-tagged RNA reporter assay, identified 22 mRNAs that are localized to bud tips. These messages encode a wide variety of proteins, including several involved in stress responses and cell wall maintenance. Many of these proteins are asymmetrically localized to buds. However, asymmetric localization also occurs in the absence of RNA transport, suggesting the existence of redundant protein localization mechanisms. In contrast to findings in metazoans, the untranslated regions are dispensable for mRNA localization in yeast. This study reveals an unanticipated widespread use of RNA transport in budding yeast.
Localization of mRNA in eukaryotic cells constitutes an important mechanism for sequestering protein activity, regulating gene expression, and establishing or maintaining cell polarity (1, 2). Studies of mRNA localization in a variety of organisms have suggested the following sequence of events: initially, an RNA molecule with specialized targeting information, or “zip code,” is recognized by a protein or protein complex that recruits a cytoskeletal motor protein (3). Next, the resulting ribonucleoprotein (RNP) complex is transported to a specific subcellular location along actin filaments or microtubules (4). Finally, the transcript becomes anchored to its final destination, where translation occurs only at the targeted location (5). In many cases, translational repression is coupled with mRNA transport to ensure that protein expression does not occur while in transit to the appropriate site of activity (5).
In Saccharomyces cerevisiae, RNA encoding the daughter cell-specific transcription factor, Ash1p, was discovered to be localized to the bud tip (6, 7). This represented the first description of RNA localization in a single-cell eukaryote. ASH1 transcripts are recognized by the She2p protein, which becomes associated with the Myo4p myosin motor via the She3p adapter protein (8–10). This RNP complex travels along actin cables to the emerging bud where the transcript is anchored and translated. The Ash1p protein then is transported into the bud nucleus, where it represses the HO locus and inhibits mating type switching. Two other proteins are also important for efficient ASH1 localization in yeast: Loc1p, a nuclear protein (11), and Khd1p, a KH (hnRNP K homology) domain containing protein that is thought to link translational repression to the localization process (12).
A second localized mRNA, IST2, was identified by a microarray-based strategy involving immunoprecipitation of She proteins, amplification of associated RNAs, and subsequent hybridization on DNA microarrays to determine their enrichment compared with those from a control immunoprecipitation (13). IST2 encodes a plasma membrane protein that is enriched in the bud. Along with bud-localized expression, Ist2p protein is prevented from diffusing into the mother cell by the septin barrier at the mother-bud junction (13).
In addition to IST2, microarray analysis identified 10 other mRNAs that were associated with the She complex (13). In situ hybridization procedures, however, revealed only IST2 and ASH1 to be asymmetrically localized, whereas the remainder were inconclusive or ambiguous (13). These same 11 transcripts were immunoprecipitated by each of She proteins independently, suggesting that these results reflect bona fide associations. However, the utility of this approach for genome-wide identification of localized mRNAs remained to be established. In this study, we have further refined various microarray approaches and developed improved methods for screening candidate transcripts for localization. With these improved methodologies, we have identified a family of messages that are localized to the tips of buds. Along with ASH1 and IST2, these bring the total number of mRNAs known to be transported by the She-protein machinery to 24.
Materials and Methods
Strains/Plasmids and Microscopy. Myc-tagged strains for microarray experiments were derived from W303 (8). Tandem affinity purification-tagged She3p and Myo4p strains were obtained from Cellzome (14). The protein A-tagged She2 strain (APG32) was generated by transforming BY4741 (15) with a PCR fragment generated from plasmid pFA6-TEVzz-kanMX6 (16), inserting two tandem IgG-binding domains downstream of SHE2 by the method of Longtine et al. (17). Strains harboring carboxyl-terminal GFP protein tags were a gift from Erin O'Shea (University of California, San Francisco) and derived from ATCC 201388. For examination of these proteins in a mutant background, SHE2 was disrupted by the method of Longtine et al. (17).
For GFP-tagging of mRNA, the pGAL–U1A plasmid was created by inserting the GAL1 promoter and four copies of the U1A aptamer site upstream of a unique NotI site and a CYC1 terminator sequence in the unique SacII site. To test sequences for localization, PCR products were amplified from genomic DNA using primers with terminal NotI sites. PCR products were cloned into the NotI site of pGAL–U1A. All constructs were confirmed by sequencing. To assess RNA localization, induction and visualization of pGAL–U1A constructs were preformed as described (8). In general, ≈50–67% of premitotic cells within a population displayed visible green RNA particles after 2 h of induction.
To quantify RNA localization, >100 premitotic cells with small to medium buds were identified and scored for GFP–RNA localized selectively to the bud versus a random distribution throughout both mother and bud. Most strains were analyzed by independent observers in a double-blind fashion.
For amino-terminal GFP tagging of proteins, pAG36 was constructed by replacing the MET25 promoter in pUG36 (unpublished data; a gift from J. H. Hegemann, Heinrich Heine University, Düsseldorf, Germany) with a GAL1 promoter via SacI/XbaI restriction sites. Genes encoding the protein to be visualized were cloned into either EcoRI or HindIII/XhoI sites. Plasmid pHS20 was constructed by Sesaki and Jensen (18) and was a gift from Michael P. Yaffe (University of California, San Diego).
To visualize proteins, strains with carboxyl-terminal GFP tags were grown to mid-log phase in yeast extract/peptone/dextrose and examined by fluorescence microscopy. To visualize amino-terminally tagged proteins, cells were grown overnight in SD–URA, diluted to 0.5 OD/ml, and induced for 1–2 h with 0.2% galactose before examination by fluorescence microscopy.
Immunoprecipitation and Microarray Analysis. Two different protocols were used in this study to identify She protein-associated RNAs (Fig. 1). First, immunoprecipitaton of She proteins, amplification of associated RNAs and hybridization to microarrays was performed as described by Takizawa et al. (13). For the second method, one liter of cells were cultured at 30°C in yeast extract/peptone/dextrose medium and collected during exponential growth by centrifugation. Cells were washed twice in 20 mM Tris·HCl, pH 8.0/140 mM KCl/1.8 mM MgCl2/0.1% Nonidet P-40/0.02 mg/ml heparin and resuspended in the above buffer containing 0.5 mM DTT, 1 mM PMSF, 0.5 μg/ml leupeptin, 0.8 μg/ml pepstatin, 20 units/ml DNase I, 100 units/ml RNasin (Promega), and 0.2 μg/ml heparin. Purification of tagged proteins and isolation of associated RNA was essentially performed as described (ref. 19 and A.P.G., P.O.B., D.H., unpublished data). Briefly, cells were broken mechanically with glass beads, and extracts were incubated with IgG-agarose beads (Sigma). The beads were washed four times, and She proteins were released from the beads by cleavage with tobacco etch virus (TEV)-protease (Invitrogen). RNA was isolated by phenol/chloroform extraction and isopropanol precipitation from TEV eluates, which corresponds to the purified fraction, and from extracts (input). Both RNA samples, input and purified, were reverse transcribed and amino-allyl labeled with the fluorescent dyes Cy3 and Cy5 (Amersham Pharmacia), respectively. The samples were mixed and competitively hybridized to yeast cDNA microarrays containing all yeast genes as described (20).
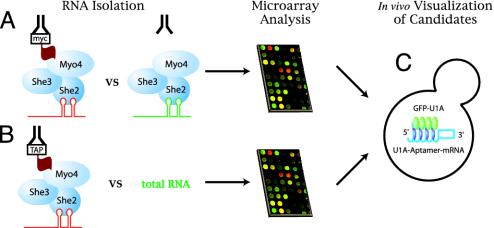
Fig. 1.
Schematic representation of microarray-based screens for localized RNAs. (A) Immunoprecipitations were performed with an anti-myc antibody from cellular extracts harboring either myc-tagged or untagged She proteins. RNAs enriched in pellets were amplified by RT-PCR and PCR, then labeled with either Cy5 (tagged) or Cy3 (untagged) nucleotides. Microarrays were probed with labeled PCR products, and enrichment for Cy5 was assessed. (B) Each She protein was protein A- or tandem affinity purification-tagged and affinity purified. Associated RNAs were labeled directly by reverse transcription with Cy5, then competitively hybridized on microarrays with total RNA from wild-type cells that had been labeled with Cy3. (C) Candidates from microarray analyses were tested for localization in vivo with a GFP–RNA tagging strategy.
Data Analysis and Retrieval. Microarray data were extracted and analyzed essentially as described (21). For further details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Results
Identification of She-Dependent Transport Candidates Using Microarray-Based Approaches. To identify mRNAs transported by the She machinery, we performed two different microarray-based experiments. First, we used the method described by Takizawa et al. (13) to immunoprecipitate myc-tagged She proteins from either tagged or untagged extracts using a monoclonal anti-myc antibody. RNAs associated with the immunoprecipitates were amplified by random-primed RT-PCR, fluorescently labeled by further PCR, and hybridized to yeast microarrays to determine which transcripts were enriched in the tagged versus untagged immunoprecipitates (method 1, Fig. 1 A).
Method 1 involved amplification of immunoprecipitated RNAs before microarray analysis. One advantage of this strategy is that rare or transiently expressed RNAs could be identified, even if initial amounts were miniscule. However, variation in the quantities of mRNA in the immunoprecipitates can lead to differential amplification during PCR. Thus, enrichment values, as estimated by intensity of signals on microarrays, are largely nonquantitative. To address this limitation, a second method was used whereby the She proteins were either protein A- or tandem affinity purification-tagged and affinity purified (14, 21). Sheassociated RNAs were then directly labeled by reverse transcription and compared with total RNA by competitive hybridization on yeast microarrays (method 2, Fig. 1B).
Within each of the two experimental methodologies, an overlapping set of transcripts were enriched in the She2p, She3p, and Myo4p immunoprecipitates, consistent with previous studies indicating that these proteins interact as a complex for mRNA transport (8–10). In addition, 13 transcripts were identified by both methodologies (see Table 2, which is published as supporting information on the PNAS web site, for details of the microarray results). Although a number of transposable elements emerged as positives, preliminary in situ hybridization analysis indicated that these RNAs are not selectively enriched in the bud (Peter Takizawa, personal communication). Thus, for the remainder of this study, we focused our efforts on only those candidates that encode predicted or known proteins.
At Least 24 mRNAs Are Transported to the Tips of Emerging Buds by the She Proteins. Of the 24 She protein-associated transcripts listed in Table 1, 11 were also identified and described by Takizawa et al. (13). However, their further studies using in situ hybridization identified only IST2 and ASH1 as localized RNAs. The remainder yielded ambiguous results because of low or variable signals, problematic background from the hybridization procedure, or poor reproducibility (13). To improve the localization assay and determine which transcripts were bona fide She-protein transport substrates, we used a U1A aptamer-based GFP tagging system described by Takizawa and Vale (8) that allows mRNA visualization by fluorescence microscopy. In this procedure, a yeast strain is transformed with two plasmids. The first expresses GFP fused to U1A, an RNA-binding protein that recognizes a specific sequence, the U1A aptamer. The second plasmid harbors a galactose-inducible promoter and four copies of the U1A aptamer fused to the 5′ end of a transcript to be analyzed. To aid in visualization, the U1A–GFP fusion carries a nuclear localization signal to direct excess, unbound protein to the nucleus. This GFP tagging procedure proved more sensitive and reproducible than in situ hybridization procedures, as expression from an inducible promoter allows overexpression and thus visualization of transient or rare transcripts that might have been difficult to detect. It is possible that U1A–GFP tagging might interfere with a transcript's localization, and overexpression might alter stoichiometry or anchoring properties of the resulting RNP complexes. However, these limitations did not detract from the utility and ease of this GFP-tagging system as a powerful secondary screen for RNA localization.
Table 1. Localized transcripts.
| RNA localization
|
|||||||
|---|---|---|---|---|---|---|---|
| Gene | Full | Coding | she2Δ | Cell cycle regulation | Predicted function | Protein localization | Tag position |
| ASH1 | Yes | Yes | No | M | Transcription | Bud nucleus (51-53) | N.C |
| BRO1 | Yes | Yes | No | None | Stress transduction | Punctae on vacuole | C |
| CLB2 | Yes | Yes | No | M | Cyclin B | Nuclei, spindle poles | C |
| CPS1 | Yes | Yes | No | None | Carboxypeptidase | Cytoplasmic punctae | N |
| DNM1 | Yes | Yes | No | S | Mitochondrial fission | Mitochondrial periphery (18, 54) | C |
| EGT2 | Yes | Partial | No | M | Cellulase | Membranes, large-bud enriched | C |
| ERG2 | Yes | Yes | No | M | Sterol isomerase | Endoplasmic reticulum | N |
| IST2 | Yes | Yes | No | None | Tranporter | Bud plasma membrane (13) | N |
| MID2 | Yes | Yes | No | None | Membrane receptor | Cell periphery, mother-bud junction | C |
| MMR1 | Yes | Yes | No | M | Unknown | Bud sites and tips, mother-bud junction | N |
| SRL1 | Yes | Yes | No | G1 | Unknown | Periphery of small buds | C |
| TPO1 | Yes | Yes | No | M | Polyamine transport | Bud plasma membrane | N |
| WSC2 | Yes | Yes | No | S | Membrane receptor | Membranes, bud-enriched | C |
| YGR046W | Yes | Yes | No | None | Unknown | Mitochondria | N |
| YJL051C | Yes | Yes | No | M | Unknown | Membranes, bud-enriched | N |
| YLR434C | Yes | Yes | No | None | Unknown | Mitochondria | N |
| YML072C | Yes | Yes | No | G2 | Unknown | Membranes, bud-enriched | C |
| YMR171C | Yes | Yes | No | None | Unknown | Endoplasmic reticulum | N |
| YNL087W | Yes | Yes | No | None | Unknown | Membranes, bud-enriched | C |
| KSS1 | Partial | ND | No | None | Mitogen-activated protein kinase | ND | ND |
| LCB1 | Partial | Partial | No | None | Endoplasmic reticulum, lipid synthesis | Endoplasmic reticulum | C |
| MET4 | Partial | Partial | No | None | Transcription | Nuclei | C |
| MTL1 | Weak | ND | No | None | MID2-like | ND | ND |
| YPL066C | Weak | ND | No | None | Unknown | ND | ND |
List of She-localized mRNAs. Also indicated are whether each transcript is cell cycle-regulated and its peak stage (49, 50), the predicted or known function, and protein localization of each. Proteins that are asymmetrically enriched in the bud are indicated by boldface. The full list of transcripts assayed for localization as well as their percentile rankings from microarray analysis can be viewed in Table 2. Full array results and additional methodology can be viewed in Supporting Materials and Methods and at http://valelab.ucsf.edu/~shepard/methods.html or http://genome-www5.stanford.edu/MicroArray/SMD/.C, carboxyl terminus; N, amino terminus. Yes, ≥90% bud localization; Partial, 50-60% localization; Weak, 15-30% localization; No, unlocalized (<5% localization; ND, not determined).
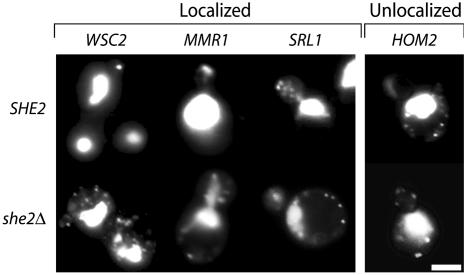
To assess localization, we used the GFP-tagging strategy to examine mRNA candidates that were identified in the top 98th percentile of one or both microarray methodologies (Table 2). To define criteria for mRNA localization, we quantified the fraction of premitotic cells with GFP–RNA concentrated in bud tips compared with those with GFP–RNA randomly distributed throughout mother and bud. Premitotic cells were chosen, because after the nuclei divide, we and others (22) observed that bud tip-localized RNAs redistribute first to the mother–daughter junction and then throughout the cytoplasm, making localization problematic to establish in larger-budded cells. We defined a “localized” mRNA as one that forms green particles that associate with the tips of small and emerging buds in >50% of premitotic cells after 1–2 h of induction, similar to what has been described for GFP-tagged ASH1 (8, 23–25). We defined an “unlocalized” mRNA as one that forms green particles that are randomly distributed between mother and bud, as has been reported for nonlocalized transcripts such as ADH1 (8, 23–25). Fig. 2 shows representative examples of these two types of distribution.
Fig. 2.
Representative examples of localized and unlocalized RNAs identified through microarray analyses. Wild-type (SHE2, Upper) or she2Δ (Lower) cells expressing GFP–RNA for indicated transcript were visualized by fluorescence microscopy. Data for all localized RNAs are shown in Fig. 5. (Bar = 2 μm.)
For the top 10 (method 1) and 9 of the top 10 (method 2) highest-ranking candidates from the array experiments (Table 2, GFP–RNA was associated with bud tips in >90% of the small-budded cells that contained visible RNA particles. In summary, of 38 tested, we found a total of 17 mRNAs that were clearly localized to bud tips, 15 that were unlocalized, and one that did not form visible GFP–RNA (SHE3). A full list of all mRNAs tested can be viewed in Table 2, and additional images of localized mRNAs can be viewed in Fig. 5, which is published as supporting information on the PNAS web site.
Although the majority of RNAs displayed clear localized or unlocalized phenotypes, several transcripts exhibited partial localization. Three of the mRNAs, MET4, LCB1, and KSS1, displayed small-bud-localized GFP–RNA in ≈50% of cells, whereas the remaining half of the cells displayed randomly distributed particles (Table 1). Two other candidates, MTL1 and YPL066w exhibited an apparently random distribution of GFP–RNA in most cells; however, 15–30% of cells displayed buds that were enriched for GFP–RNA, something that was never observed in “unlocalized” candidates. This variability in localization efficiency might result from attenuation of anchoring, translational, or localization signals in the transcripts caused by an artifact of the U1A–GFP tagging strategy. Alternatively, it might reflect a genuine difference in affinity of these transcripts for the She machinery in vivo. Because of their weak localization activity, we chose not to pursue MTL1 and YPL066W for further analysis.
To determine whether bud-specific localization of mRNA requires the She protein machinery, we examined mRNA localization in she2Δ cells. As described previously for ASH1 and IST2, all of the localized RNAs (Table 1), as well as the five partially localized mRNAs described above, were delocalized from bud tips and appeared randomly distributed throughout both mothers and buds (Fig. 2). Similar delocalization was observed in myo4Δ or she3Δ mutants for three transcripts that were tested: MID2, IST2, and TPO1 (data not shown). These data indicate that, like ASH1 and IST2, all of the new localized transcripts require the She protein complex to localize to the tips of growing buds.
Many, But Not All, Proteins Encoded by Localized mRNAs Show Asymmetric Distributions. mRNA localization is essential for the asymmetric distribution of Ash1p protein to bud nuclei and Ist2p protein to the bud plasma membrane (6, 7, 13). To determine the normal localization of proteins encoded by the RNA candidates, we tagged each protein with GFP and assessed its subcellular distribution by fluorescence microscopy (Table 1). Because ASH1 and most localized RNAs from metazoan cells have been shown to contain important RNA localization signals in or near their 3′ UTRs (1, 6, 7), we initially tagged most of the proteins at the amino terminus and placed their expression under control of the inducible GAL1 promoter. By this strategy, we also hoped to see expression of rare or transient transcripts that might have been difficult to detect at endogenous levels.
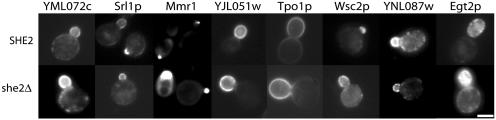
GFP-tagged proteins Tpo1p, YJL051p, and Srl1p were localized specifically to the periphery of buds, similar to what was observed for Ist2p (13) (Fig. 3). However, some of the proteins, including several that were predicted to be membrane associated, such as Mid2p and Wsc2p, appeared as amorphous green particles in the cytoplasm. To determine whether this localization was genuine or perhaps an artifact of overexpression or tag position, we obtained strains harboring versions of the proteins that were chromosomally tagged with GFP at their carboxyl termini and expressed from their endogenous promoters. Many of these GFP-tagged proteins showed asymmetric localization to the bud (Fig. 3), including Srl1p, YJL051p, YML072p, YNL087p, and Wsc2p, whereas GFP-Mmr1p (YLR190w) localized to discrete punctae at future bud sites, small buds, and mother-bud junctions in postmitotic cells. The remaining proteins were segregated symmetrically to various subcellular destinations as indicated in Table 1. Images for these “symmetrically distributed” proteins can be viewed in Fig. 6, which is published as supporting information on the PNAS web site.
Fig. 3.
Asymmetric protein localization is independent of She-based RNA transport. Wild-type (SHE2, Upper) or she2Δ (Lower) cells expressing GFP-tagged versions of the indicated proteins were visualized by fluorescence microscopy. Full results are listed in Table 1. (Bar = 2 μm.)
Asymmetric Protein Localization Can Be Achieved in the Absence of mRNA Localization. She-dependent RNA sorting is essential for asymmetric localization of Ash1p and Ist2p to the bud nucleus and bud membrane, respectively (6, 7, 13). To determine whether RNA transport to the bud tip is essential for normal protein distribution, we expressed each of the GFP-tagged proteins in she2Δ cells and examined their distributions in vivo. Surprisingly, protein localizations, including those that were asymmetric, were unaltered in she2Δ mutants (Fig. 3). Thus, unlike Ash1p and Ist2p (6, 7, 13), the majority of the She protein-RNA transport substrates encode proteins with redundant targeting information such that they are distributed correctly in the absence of RNA transport.
The Coding Regions of RNA Transport Substrates Are Sufficient for Targeting to the Bud. Zip codes are regions of RNA sequence that are sufficient to confer localization of a reporter RNA (1). In metazoans, all published zip codes to date have mapped to 3′ or (rarely) to 5′ UTRs of a transcript (1). However, the yeast ASH1 mRNA contains zip codes in both the coding sequence and 3′ UTR (6, 7, 24, 26). To determine whether the other Shetransported mRNAs contain zip codes, we created U1A–GFP-tagged versions of each localized message that included only the coding sequence and the CYC1 terminator rather than endogenous termination sequences. Surprisingly, the coding sequence alone was sufficient to enable transport to the tips of growing buds for the all of the localized mRNAs (Table 1). In most cases, the efficiency of localization was similar to that observed for the full-length constructs (Fig. 4 and Fig. 7, which is published as supporting information on the PNAS web site). However, the EGT2 coding transcript conferred bud localization in only 15–20% of the cells. For 12 of the localized mRNAs, we assessed the localization of the 3′ UTR alone (here defined as the 500-bp region immediately downstream of the stop codon). The 3′ UTRs of ERG2, CLB2, and EGT2 conferred partial localization in the U1A–GFP reporter system, whereas the other tested 3′ UTRs appeared unlocalized (IST2, YGR046w, MMR1, TPO1, YML072c, SRL1, MID2, YMR171c) or did not form detectable GFP–RNA (WSC2) (data not shown). These data indicate that zip code information within the coding region is usually sufficient for the recognition and transport of mRNA by the She protein machinery.
Fig. 4.
Coding sequences of She-transport substrates are largely sufficient for RNA localization. Wild-type cells expressing GFP–RNA for coding regions of indicated transcripts were visualized by fluorescence microscopy. Three representative transcripts are shown. Additional localized coding sequences are shown in Fig. 7, and a summary of the data is presented in Table 1. (Bar = 2 μm.)
Discussion
We have identified 22 previously undescribed substrates for the yeast She-protein RNA transport machinery through a combination of protein immunoprecipitation, DNA microarray analysis, and GFP–RNA visualization. The GFP–RNA tagging strategy, as opposed to in situ hybridization, proved crucial for documenting the localization of additional RNAs compared with the original study by Takizawa et al. (13). This study reveals that RNA localization in yeast is more widespread than previously appreciated. It is possible that these recently identified mRNAs represent the majority of the She-protein cargoes, given that the lower-ranking candidates in the screen were less likely to localize to bud tips. However, our experiments were performed on cells that were growing exponentially in rich media. Any transcripts that are expressed only transiently or under a limited set of circumstances would likely have been overlooked. In addition, some weakly binding RNAs may dissociate from the She complex in vitro before immunoprecipitation. Finally, it is possible that alternative RNA localization mechanisms exist in yeast that are independent of She function. Indeed, RNA targeting to the mitochondria has been described in budding yeast (27). Although the mechanism for this process has yet to be established, none of the mitochondrial-targeted mRNAs were identified in our microarray experiments as substrates for the She machinery.
Predicted Functions of Proteins Encoded by Localized mRNAs. The proteins encoded by localized mRNAs are diverse, although some appear to participate in common pathways related to sensing or responding to stress. WSC2 encodes a heat shock sensor that transduces signals via a MPK1 pathway, whereas MID2 transmits a similar signal in response to α-factor. Mutations in either of these genes can lead to osmotic sensitivity (28, 29), and a mid2Δ mutation is suppressed by overexpression of Wsc2p (29). MTL1 is thought to encode a cell wall sensor and displays significant sequence homology with both WSC2 and MID2 (29, 30). Two other localized RNAs, BRO1 and KSS1, encode components of mitogen-activated protein kinase signaling pathways that regulate cellular responses to various environmental stresses (31, 32), whereas IST2 and TPO1 encode membrane transporters that may modulate intracellular ion concentrations (33). The functional significance of this potentially interconnected group of localized messages requires further elucidation. However, there may exist interactions or interdependencies among She substrates at the RNA or protein level that do not readily reveal themselves through traditional genetic methods.
Several localized RNAs encode proteins that play roles in synthesis and modification of the plasma membrane and cell wall. ERG2 and LCB1 encode enzymes involved in lipid synthesis (34), whereas EGT2 encodes a cellulase that may be involved in cell separation (35–37). Localization of these proteins to the bud would concentrate their activities at the most active site of cell growth and remodeling.
Ten of the twenty-four She-localized mRNAs are transcribed from cell-cycle-regulated genes (Table 1), a significantly higher proportion than the genomic representation of such genes (10%) (www.yeastgenome.org). Seven of these transcripts have peak expressions at M or M/G1 (Table 1). She2 transcription also peaks at M phase, suggesting that RNA transport machinery might be maximally expressed at the time of its greatest usage (www.yeastgenome.org). Of the other M-phase regulated transcripts, CLB2 is of particular interest, because the mRNAs for cyclin B homologs are localized in a variety of metazoan organisms (1). Although we only were able to detect Clb2p in nuclei and spindle pole bodies, domain analysis has revealed that there might be a subpopulation of Clb2p present at the bud tip and mother-bud junction (38). A bud-specific pool of Clb2p might be important for initiating a daughter-specific genetic program. Perhaps the She machinery is responsible for localizing a subset of Clb2 that resides outside the nucleus and is expressed only transiently or at levels that are difficult to detect by conventional means. Indeed, it is possible that asymmetric subpopulations could exist for other proteins encoded by localized messages whose overall distributions appeared random in our visualization assay.
Many of the localized RNAs encode proteins that are known or predicted to associate with membranes. Several of these are asymmetrically enriched in the bud, and many encode proteins of unknown function (e.g., YML072c, YJL051c, YNL087w). Others (YMR046w, YLR434c, DNM1, ERG2, LCB, MET4, CLB2) encode proteins that are symmetrically distributed to subcellular membranous structures such as mitochondria, endoplasmic reticulum, or nuclei. Despite their apparent symmetry, it is possible that these proteins have asymmetric distributions that were disrupted by introduction of GFP tags and/or overexpression. Alternatively, these proteins may be synthesized at the site of the localized transcript, but the recently translated proteins may rapidly equilibrate between mother and bud, thereby creating a symmetric steady-state distribution.
The Role of RNA Transport. mRNA transport is generally thought to play a primary role in creating an asymmetric protein distribution in yeast (6, 7). However, other than ASH1, there is no evidence to suggest that asymmetry is important for the function of proteins encoded by localized transcripts. Indeed, the abolition of RNA transport by deletion of the She machinery confers no obvious defects on growth or fitness (39). However, we found that most asymmetrically distributed proteins encoded by RNA transport substrates maintained their localization in the absence of RNA transport. Thus, these proteins must themselves contain targeting information that allows for their appropriate posttranslational segregation. For the plasma membrane proteins, this redundancy may involve the secretory pathway, which includes actin-based transport of vesicles to the bud. The ability of proteins encoded by localized mRNAs to sort to their appropriate locations posttranslationally is consistent with observations in other organisms (40). For example, both Prospero mRNA and protein are asymmetrically localized in Drosophila neuroblasts, yet the protein can achieve asymmetry even when its RNA is symmetrically dispersed (41). These results might imply the necessity of alternative targeting mechanisms for important biological functions.
The observation that several She substrates encode proteins that are not asymmetrically distributed raises the possibility that She proteins and mRNA transport might also serve functions that are not directly related to protein localization. One possibility is that the She complex may provide buds with a “start-up package” of mRNA, so that a daughter cell can respond to stimuli without initiating a round of its own transcription. This notion is analogous to the transfer of maternal mRNAs into developing Drosophila oocytes. Such a feature might give fitness advantages to progeny under certain environmental stresses, perhaps some that are too subtle to have been detected by standard competitive growth assays. Alternatively, She proteins might affect the anchoring or translation of their substrates, either by direct interaction with RNA or through interactions with accessory molecules, such as Khd1 (11) or Loc1 (12). Studies in Drosophila have identified several molecules that integrate RNA transport with related processes such as splicing, nuclear export, and translational control (42, 43). It is likely that similar relationships in yeast will emerge upon further elucidation of the functional contribution of each component of the She mRNA transport complex.
The Coding Region Contains mRNA Localization Elements. The identification of 24 substrates for the She machinery provides new resources for identifying sequences that are important for recognition by She2p. Determination of zip code sequences in yeast as well as higher organisms has been challenging, because this information is thought to reside in secondary or tertiary structures that are difficult to predict. In contrast to other organisms such as Drosophila, where multiple RNA localization pathways exist and few zip code-binding adapter molecules have been identified, these yeast RNAs interact with the same adapter molecule, She2p. In this study, we have determined that the majority of yeast zip code domains lie within the coding sequences of the genes. A recent study from Chartrand et al. (44) reported that localization elements in the ASH1 coding sequence serve to decelerate translation, presumably so that ASH1 mRNA is not prematurely expressed while in transit to the bud tip. The number and position of zip code elements in the coding sequences of other localized yeast mRNAs may prove similarly important for timing of protein expression. Indeed, it is even possible that RNA localization to the bud tip might be a secondary consequence of translational regulation by certain zip code elements, which could explain why several of the proteins encoded by localized RNAs are not found specifically in the bud.
With this repertoire of localized RNAs, mapping of zip code motifs and analysis of their contribution to protein expression will yield insight into how these sequences mediate both motility and translation. In addition, functional and computational analyses of these regions could aid in the development of predictive tools for zip code identification in yeast as well as other organisms.
Applications in Other Systems. We have demonstrated that a microarray approach based on purification of RNA-binding proteins, combined with a robust reporter system as a secondary screen, provides a powerful technique for identifying localized RNAs. This strategy could be used to discover new RNA cargoes in a variety of organisms. Any molecule that is known or suspected to play a role in transport, whether it is a motor protein or an adapter, could be used as a handle to purify and identify associated RNAs. An aptamer–GFP-binding strategy has now been described that allows visualization of RNAs in mammalian cells (45), but other RNA visualization techniques, such as in situ hybridization (13, 46) and microinjection (47, 48), also can be used as secondary screens. Given the relative ease of these procedures and the increasing availability of genomic tools, it is possible that analogous experiments will lead to the rapid identification of large repertoires of localized RNAs in a wide variety of organisms.
Supplementary Material
Acknowledgments
We gratefully acknowledge Maki Inada for assistance with microarray analysis and Alan Kutach for helpful advice on the manuscript and figures. We also thank Drs. P. Preker, J. H. Hegemann, and M. P. Yaffe for plasmids, and Dr. Michael Rosbash for initial advice on U1A–GFP tagging. This work was supported in part by a grant from the Jane Coffin Childs Memorial Fund for Medical Research (to K.A.S.), a grant from the Human Frontier Science Program (to A.P.G.), a National Science Foundation fellowship (to A.J.) and National Institutes of Health Grant 38499 (to R.D.V.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: RNP, ribonucleoprotein.
References
- 1.Bashirullah, A., Cooperstock, R. L. & Lipshitz, H. D. (1998) Annu. Rev. Biochem. 67, 335–394. [DOI] [PubMed] [Google Scholar]
- 2.Tekotte, H. & Davis, I. (2002) Trends Genet. 18, 636–642. [DOI] [PubMed] [Google Scholar]
- 3.Kislauskis, E. H. & Singer, R. H. (1992) Curr. Opin. Cell Biol. 4, 975–978. [DOI] [PubMed] [Google Scholar]
- 4.Oleynikov, Y. & Singer, R. H. (1998) Trends Cell Biol. 8, 381–383. [DOI] [PubMed] [Google Scholar]
- 5.Johnstone, O. & Lasko, P. (2001) Annu. Rev. Genet. 35, 365–406. [DOI] [PubMed] [Google Scholar]
- 6.Long, R. M., Singer, R. H., Meng, X., Gonzalez, I., Nasmyth, K. & Jansen, R. P. (1997) Science 277, 383–387. [DOI] [PubMed] [Google Scholar]
- 7.Takizawa, P. A., Sil, A., Swedlow, J. R., Herskowitz, I. & Vale, R. D. (1997) Nature 389, 90–93. [DOI] [PubMed] [Google Scholar]
- 8.Takizawa, P. A. & Vale, R. D. (2000) Proc. Natl. Acad. Sci. USA 97, 5273–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long, R. M., Gu, W., Lorimer, E., Singer, R. H. & Chartrand, P. (2000) EMBO J. 19, 6592–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohl, F., Cruse, K., Frank, A., Ferring, D. & Jansen, R. P. (2000) EMBO J. 19, 5514–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long, R. M., Gu, W., Meng, X., Gonsalvez, G., Singer, R. H. & Chartrand, P. (2001) J. Cell Biol. 153, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irie, K., Tadauchi, T., Takizawa, P. A., Vale, R. D., Matsumoto, K. & Herskowitz, I. (2002) EMBO J. 21, 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takizawa, P. A., DeRisi, J. L., Wilhelm, J. E. & Vale, R. D. (2000) Science 290, 341–344. [DOI] [PubMed] [Google Scholar]
- 14.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 15.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P. & Boeke, J. D. (1998) Yeast 14, 115–132. [DOI] [PubMed] [Google Scholar]
- 16.Preker, P. J., Kim, K. S. & Guthrie, C. (2002) RNA 8, 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. & Pringle, J. R. (1998) Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 18.Sesaki, H. & Jensen, R. E. (1999) J. Cell Biol. 147, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M. & Seraphin, B. (1999) Nat. Biotechnol. 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- 20.DeRisi, J. L., Iyer, V. R. & Brown, P. O. (1997) Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 21.Lieb, J. D., Liu, X., Botstein, D. & Brown, P. O. (2001) Nat. Genet. 28, 327–334. [DOI] [PubMed] [Google Scholar]
- 22.Beach, D. L., Salmon, E. D. & Bloom, K. (1999) Curr. Biol. 9, 569–578. [DOI] [PubMed] [Google Scholar]
- 23.Bertrand, E., Chartrand, P., Schaefer, M., Shenoy, S. M., Singer, R. H. & Long, R. M. (1998) Mol. Cell 2, 437–445. [DOI] [PubMed] [Google Scholar]
- 24.Chartrand, P., Meng, X. H., Singer, R. H. & Long, R. M. (1999) Curr. Biol. 9, 333–336. [DOI] [PubMed] [Google Scholar]
- 25.Bloom, K. S., Beach, D. L., Maddox, P., Shaw, S. L., Yeh, E. & Salmon, E. D. (1999) Methods Cell Biol. 61, 369–383. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez, I., Buonomo, S. B., Nasmyth, K. & von Ahsen, U. (1999) Curr. Biol. 9, 337–340. [DOI] [PubMed] [Google Scholar]
- 27.Corral-Debrinski, M., Blugeon, C. & Jacq, C. (2000) Mol. Cell. Biol. 20, 7881–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zu, T., Verna, J. & Ballester, R. (2001) Mol. Genet. Genomics 266, 142–155. [DOI] [PubMed] [Google Scholar]
- 29.Ketela, T., Green, R. & Bussey, H. (1999) J. Bacteriol. 181, 3330–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajavel, M., Philip, B., Buehrer, B. M., Errede, B. & Levin, D. E. (1999) Mol. Cell. Biol. 19, 3969–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickas, M. E. & Yaffe, M. P. (1996) Mol. Cell. Biol. 16, 2585–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courchesne, W. E., Kunisawa, R. & Thorner, J. (1989) Cell 58, 1107–1119. [DOI] [PubMed] [Google Scholar]
- 33.Tomitori, H., Kashiwagi, K., Sakata, K., Kakinuma, Y. & Igarashi, K. (1999) J. Biol. Chem. 274, 3265–3267. [DOI] [PubMed] [Google Scholar]
- 34.Ashman, W. H., Barbuch, R. J., Ulbright, C. E., Jarrett, H. W. & Bard, M. (1991) Lipids 26, 628–632. [DOI] [PubMed] [Google Scholar]
- 35.Kovacech, B., Nasmyth, K. & Schuster, T. (1996) Mol. Cell. Biol. 16, 3264–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan, X. & Heitman, J. (2000) Mol. Cell. Biol. 20, 8364–8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colman-Lerner, A., Chin, T. E. & Brent, R. (2001) Cell 107, 739–750. [DOI] [PubMed] [Google Scholar]
- 38.Hood, J. K., Hwang, W. W. & Silver, P. A. (2001) J. Cell Sci. 114, 589–597. [DOI] [PubMed] [Google Scholar]
- 39.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 40.Serano, T. L. & Cohen, R. S. (1995) Development (Cambridge, U.K.) 121, 3013–3021. [DOI] [PubMed] [Google Scholar]
- 41.Broadus, J., Fuerstenberg, S. & Doe, C. Q. (1998) Nature 391, 792–795. [DOI] [PubMed] [Google Scholar]
- 42.Mansfield, J. H., Wilhelm, J. E. & Hazelrigg, T. (2002) Development (Cambridge, U.K.) 129, 197–209. [DOI] [PubMed] [Google Scholar]
- 43.Dreyfuss, G., Kim, V. N. & Kataoka, N. (2002) Nat. Rev. Mol. Cell Biol. 3, 195–205. [DOI] [PubMed] [Google Scholar]
- 44.Chartrand, P., Meng, X. H., Huttelmaier, S., Donato, D. & Singer, R. H. (2002) Mol Cell 10, 1319–1330. [DOI] [PubMed] [Google Scholar]
- 45.Fusco, D., Accornero, N., Lavoie, B., Shenoy, S. M., Blanchard, J. M., Singer, R. H. & Bertrand, E. (2003) Curr. Biol. 13, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Femino, A. M., Fay, F. S., Fogarty, K. & Singer, R. H. (1998) Science 280, 585–590. [DOI] [PubMed] [Google Scholar]
- 47.Pederson, T. (2001) Nucleic Acids Res. 29, 1013–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkie, G. S. & Davis, I. (2001) Cell 105, 209–219. [DOI] [PubMed] [Google Scholar]
- 49.Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho, R. J., Campbell, M. J., Winzeler, E. A., Steinmetz, L., Conway, A., Wodicka, L., Wolfsberg, T. G., Gabrielian, A. E., Landsman, D., Lockhart, D. J. & Davis, R. W. (1998) Mol. Cell 2, 65–73. [DOI] [PubMed] [Google Scholar]
- 51.Sil, A. & Herskowitz, I. (1996) Cell 84, 711–722. [DOI] [PubMed] [Google Scholar]
- 52.Bobola, N., Jansen, R. P., Shin, T. H. & Nasmyth, K. (1996) Cell 84, 699–709. [DOI] [PubMed] [Google Scholar]
- 53.Jansen, R. P., Dowzer, C., Michaelis, C., Galova, M. & Nasmyth, K. (1996) Cell 84, 687–697. [DOI] [PubMed] [Google Scholar]
- 54.Bleazard, W., McCaffery, J. M., King, E. J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J. & Shaw, J. M. (1999) Nat. Cell Biol. 1, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.