Abstract
The initial interaction of human cytomegalovirus with fibroblasts triggers, and then partially blocks, an innate immune response pathway that leads to the induction of IFN-responsive genes and proinflammatory chemokines. Infection of fibroblasts with human cytomegalovirus inhibited their ability to respond to exogenous IFN. Consistent with the observation that the block did not depend on de novo viral protein synthesis, ectopic expression of the viral UL83-coded pp65, an abundant virion protein, inhibited IFN signaling. Furthermore, DNA array analysis showed that infection with a pp65-deficient mutant virus caused a much stronger induction of many IFN-response and proinflammatory chemokine RNAs than infection with wild-type virus. The nuclear DNA-binding activities of transcription factors NF-κB and IRF1 were induced to a much greater extent after infection with the pp65-deficient mutant than with wild-type virus. IFN-stimulated gene factor 3 DNA-binding was modestly enhanced, whereas IRF3 activity was not affected by mutation of pp65. Together, these results imply that pp65, which is delivered to newly infected cells in the virion, antagonizes a pathway that affects NF-κB and IRF1 and prevents the accumulation of mRNAs encoded by numerous cellular antiviral genes.
Human cytomegalovirus (HCMV) is a ubiquitous β-herpes virus, and is a significant cause of disease for unborn children and immuno-compromised patients. Infections begin at mucosal epithelial surfaces before spreading to a variety of other cell-types and tissues. HCMV infection provokes a potent T cell response that suppresses the infection, but, like other herpesviruses, a lifelong latent infection is established after the primary infection is cleared. Latent virus is found in monocytes and CD34+ hematopoietic stem cells (1–3). HCMV executes multiple immune-evasive activities in infected cells. The viral proteins US2, US3, US6, and US11 cooperate to inhibit MHC-I presentation of viral peptides to CD8 T cells (4, 5). The secreted HCMV protein UL21.5 acts as a soluble decoy receptor for the proinflammatory chemokine RANTES (D. Wang, W. Bresnahan, and T.S., unpublished data), and HCMV expresses a viral homologue of IL-10, an antiinflammatory inter-leukin (6).
A key component of the innate-immune response to viral infections is the IFN pathway. IFNs are cytokines that are synthesized in response to virus infection. They bind to their cognate receptors on target cells and activate a signaling pathway that coordinately induces a set of IFN-responsive genes, many of which exhibit antiviral activity. Fibroblasts, which are commonly used to study HCMV replication, produce primarily IFN-β. HCMV infection triggers the accumulation of many IFN-responsive mRNAs (7–10). However, we have previously shown that a substantially greater number of IFN-responsive genes are induced at 6 h after infection with HCMV particles that were inactivated by UV treatment before infection than with replication-competent virus (10). This implies that, immediately after infection, HCMV synthesizes a gene product that suppresses the up-regulation of IFN-responsive genes. However, UV-inactivation of viral particles did not lead to full activation of the IFN pathway, possibly indicating that another block is instituted that is independent of de novo HCMV gene expression in newly infected cells.
Here we demonstrate that the abundant HCMV virion protein, pp65, blocks the induction of some, but not all, IFN-responsive genes by inhibiting an innate immune response pathway that leads to the activation of NF-κB and IRF1.
Materials and Methods
Cells, Viruses, and Reagents. Primary human foreskin fibroblasts at passage 9–17 were maintained in medium supplemented with 10% FCS. A plaque-purified derivative of the AD169 strain of HCMV, originally obtained from the American Type Culture Collection, was used as the wild-type virus in these studies. The pp65-deficient mutant, HCMVΔpp65, contained the bacterial neomycin phosphotransferase gene in place of the AD169 UL83 ORF (11). Virus particles were partially purified from cell culture medium by centrifugation through a sorbitol cushion and resuspended in PBS, stored as virus stocks at –80°C, and stocks were titered by plaque assay on human fibroblasts. In all experiments, fibroblasts were infected at a multiplicity of 5 plaque-forming units per cell for 1 h before conditioned medium was added back to the dish.
Recombinant IFN-α was from Calbiochem, and all antibodies were obtained from Santa Cruz Biotechnology except antibody to p-stat1 (Cell Signaling Technology) and p-stat2 (Upstate Biotechnology).
DNA Array Analysis. Infected fibroblasts were suspended in TRIzol and then stored at –80°C until they were processed by purification of total RNA. A total of 5 μg of each RNA sample was used as a template for cDNA synthesis in a reaction that was primed with an oligonucleotide containing an oligo(dT) stretch and a T7 RNA polymerase promoter. The cDNA was used to make biotin-labeled cRNA probes with an RNA transcript labeling kit (Enzo Diagnostics). The cRNA was purified to remove unincorporated ribonucleotides, and 15 μg was fragmented at 95°C for 30 min in buffer containing 40 mM Tris-acetate (pH 8.1), 100 mM KOAc, and 30 mM KOAc. The fragmented cRNA was hybridized to HG-U95Av2 arrays for 16 h at 45°C. The arrays were washed and stained as described by using the Affymetrix antibody amplification protocol (12). Scanning was performed by using an Agilent gene chip scanner. Scanned chip data sets were analyzed by using Affymetrix GeneChip analysis software. The “fold change” in intensity for the probe set was averaged for two replicate experiments.
Electrophoretic Mobility Shift Assay (EMSA). Nuclear extracts were prepared and used for EMSAs following the protocol described previously (13). The sequences for the oligonucleotides used as probes are as follow (5′–3′): IFN-stimulated response element (ISRE) (sense), GATCGGGAAAGGGAAACCGAAACTGAAGCCA; ISRE (antisense), TGGCTTCAGTTTCGGTTTCCCTTTCCCGATC; NF-κB (sense), AGTTGAGGGGACTTTCCCAGGC; NF-κB (antisense), GCCTGGGAAAGTCCCCTCAACT; NF-κB (mutant sense), AGTTGAGGCGACTTTCCCAGG; NF-κB (mutant antisense), GCCTGGGAAAGTCGCCTCAACT. A total of 5 μg of nuclear protein was used per binding reaction.
Immunofluorescence. Fibroblasts growing on glass coverslips were processed for immunofluorescence at room temperature. Cells were washed with PBS, fixed with 2% paraformaldehyde for 15 min, washed again, permeabilized with 0.1% Triton X-100, and blocked with 2% BSA in PBS containing 0.05% Tween 20 for 60 min. Cells were next incubated with primary antibody (diluted 1:40 in blocking buffer) for 60 min, and then with fluorescently labeled secondary anti-mouse or anti-rabbit antibody (1:1,000 dilution in blocking buffer) plus 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 60 min. The coverslips were then washed and mounted on a slide with 17 μl of SlowFade (Molecular Probes) before being analyzed by confocal microscopy.
Results
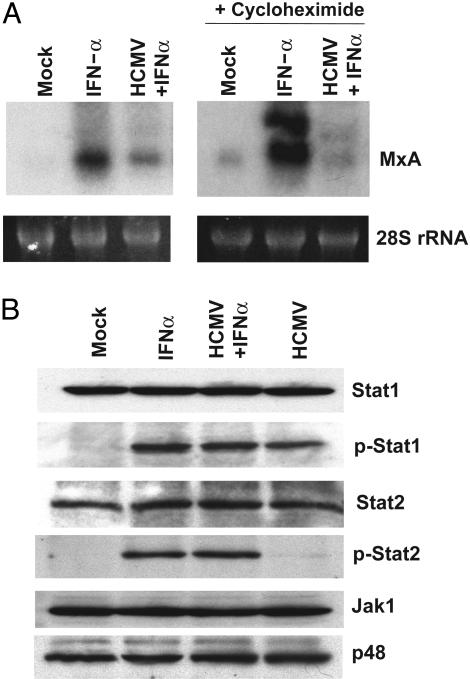
HCMV Blocks the Ability of Infected Cells to Respond to Exogenously Added IFN. To investigate how HCMV infection affects the IFN response pathway, we treated fibroblasts with IFN-α at 4 h after mock infection or infection with wild-type virus. The cells were harvested 2 h later, RNA was prepared and tested for the induction of the MxA IFN-responsive gene (Fig. 1A Left). The level of MxA RNA was consistently lower in infected than in mock-infected fibrobasts, implying that HCMV infection antagonizes the IFN response pathway. The block to IFN signaling was even more apparent when the experiment was carried out in the presence of cycloheximide (Fig. 1A Right). This finding indicates that the ability of HCMV to block the induction of MxA RNA does not depend on de novo viral protein synthesis and that a structural component of the virus particle is responsible. The finding that the block is more pronounced in the presence of cycloheximide is likely due to the fact that the drug prevents the production of IFN, blocking its ability to contribute to the induction of MxA RNA. Curiously, the MxA RNA appears as a doublet in the presence of cycloheximide. The doublet was consistently observed in multiple experiments, and the reason for the generation of two bands is unclear.
Fig. 1.
HCMV blocks IFN signaling. (A) Untreated fibroblasts (Left) or fibroblasts continuously treated with 100 μg/ml cycloheximide beginning 1 h before infection (Right) were mock-infected or infected with wild-type HCMV. At 4 h after infection, IFN-α (2,000 units/ml) was added to the cultures, and at 6 h after infection, RNA was harvested and assayed for the IFN-sensitive gene MxA by Northern blot. The level of 28S ribosomal RNA (rRNA) was monitored by ethidium bromide fluorescence as a loading control. (B) Fibroblasts were pretreated with 100 μg/ml cycloheximide beginning 1 h before mock-infection or infection with wild-type HCMV. At 4 h after infection, 2,000 units/ml IFN-α was added to infected and mock-infected cells. At 6 h after infection, cells were lysed and extracts were analyzed by Western blot assay for the indicated proteins by using specific antibodies. Stat1 and Stat2 designate total Stat protein; p-Stat1 and p-Stat2 designate phosphorylated Stat proteins.
Treatment of cells with IFN-α or -β results in activation of the Janus kinase 1 (Jak1) and Tyk2 kinases, which promote the phosphorylation of the signal transducer and activator of transcription (Stat)1 and -2 proteins. The phosphorylated Stat proteins then form a heterodimeric complex that translocates to the nucleus and associates with p48 to form the IFN-stimulated gene factor 3 (ISGF3) complex. This transcription factor up-regulates the expression of IFN-response genes by binding to the ISRE promoter element (14, 15). It has previously been reported that HCMV infection blocks IFN signaling at much later times after infection (24–48 h) than we have observed here, and that this block correlates with loss of Jak1 and p48 (16). Western blot analyses (Fig. 1B) demonstrated that the levels of Jak1, Stat1, Stat2, and p48 were not affected by HCMV at 6 h after infection, when the virus has clearly blocked the induction of MxA RNA. HCMV infection induced Stat1 phosphorylation to the same extent as treatment with IFN-α. Little Stat-2 phosphorylation was caused by HCMV infection in the presence of cycloheximide, although Stat-2 phosphorylation could still be strongly induced in infected cells by the addition of IFN-α. We therefore conclude that HCMV prevents the infected cell from inducing MxA RNA in response to exogenously added IFN at 6 h after infection by a mechanism other than the failure to phosphorylate Stat proteins or to modulate Jak1 or p48 levels.
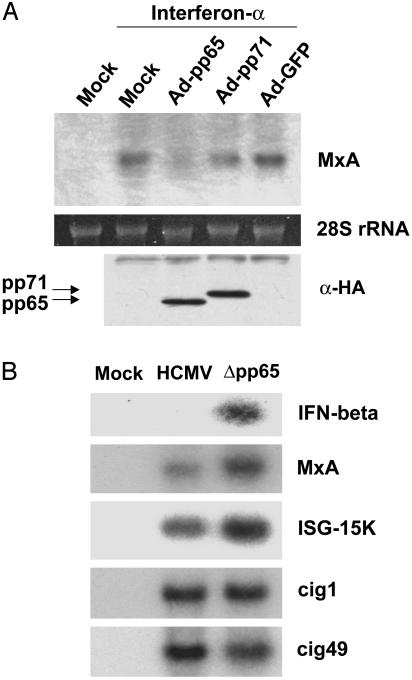
IFN-α Signaling Is Blocked by pp65. To search for a virion component that can block the induction of MxA RNA in response to IFN, we initially tested the activity of pp65 and pp71, two abundant virus-coded proteins that are packaged into the tegument domain of the HCMV particle. We infected fibroblasts with recombinant adenoviruses that lack the adenovirus E1A gene and express either pp65, pp71, or GFP. At 24 h after infection, we added IFN-α to infected cells for 1 h and monitored the induction of MxA RNA by Northern blot assay (Fig. 2A). Cells expressing pp65 accumulated much less MxA RNA than cells expressing pp71 or GFP, indicating that pp65 can block elements of the IFN signaling pathway. To confirm and extend this result, the accumulation of RNAs encoded by several additional IFN-responsive genes was monitored by Northern blot assay after infection with wild-type HCMV or a mutant (Δpp65) lacking the UL83 ORF, which codes for pp65 (Fig. 2B). At 6 h after infection, some RNAs (IFN-β, MxA, ISG-15K) were more strongly induced by Δpp65 than by wild-type HCMV, indicating that pp65 inhibits their induction in HCMV-infected cells. Other IFN response genes (cig1, cig49), however, showed little difference in their response to the two viruses. This finding implies that pp65 plays a role in suppressing a distinct subset of IFN response genes. This result is reminiscent of the finding that UV inactivation of the virus particles leads to a much stronger induction of some IFN-responsive genes than others (10).
Fig. 2.
IFN-α signaling is antagonized by pp65. (A) Fibroblasts were infected with recombinant adenoviruses expressing hemagglutinin (HA)-tagged pp65 (Ad-pp65), HA-tagged pp71 (Ad-pp71), or GFP (Ad-GFP) at a multiplicity of 104 particles per cell. At 24 h after infection, 2,000 units/ml IFN-α was added for 1 h. RNA was then prepared, and the level of MxA mRNA was assayed by Northern blot (Upper). Protein samples also were assayed by using an antibody specific for the HA epitope (α-HA) (Lower) to confirm the expression of pp65 and pp71. (B) Fibroblasts were infected with wild-type HCMV or a pp65-deficient mutant (Δpp65). After 6 h, cells were harvested and RNA was extracted and analyzed by Northern blot using 32P-labeled probes specific for the indicated IFN-responsive genes.
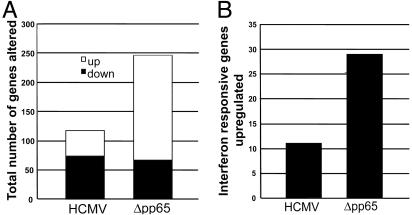
Global Effect of pp65 on Cellular Gene Expression. We next examined the effect of pp65 on the accumulation of a larger set of cellular RNAs. Fibroblasts were infected with wild-type HCMV or Δpp65 for 6 h, and cellular RNA levels were analyzed by using the Affymetrix HG-U95A array that monitors ≈12,600 cellular transcripts. As shown in Fig. 3A, at this time after infection, wild-type HCMV altered the level of 117 cellular RNAs by a factor of ≥3 (43 up-regulated and 74 down-regulated), whereas the pp65-deficient mutant altered the expression of 246 cellular RNAs (179 upregulated and 67 down-regulated). This result demonstrates that pp65 is required to suppress the induction of numerous cellular RNAs early in infection.
Fig. 3.
Global effect of pp65 on cellular gene expression. DNA array analysis was carried out on duplicate samples of RNA prepared from fibroblasts at 6 h after infection with wild-type HCMV or Δpp65. (A) The total number of RNAs up- or down-regulated by a factor of ≥3 in two replicates was determined and graphed. (B) The number of RNAs induced by a factor ≥3 in both replicates, which were previously shown to be induced by IFN (10), was determined and graphed.
Many cellular RNAs (n = 180) were elevated to a greater extent (≥2-fold) in response to Δpp65 than to the wild-type virus. This group of RNAs included many known IFN-responsive genes (Table 1) and proinflammatory chemokines (Table 2), including IFN-β itself. We previously defined a set of 79 genes that are induced by a factor of ≥3 by treatment of fibroblasts with IFN-α (10). The pp65-deficient mutant up-regulated 29 of these IFN response genes by a factor of ≥3 at 6 h after infection. By contrast, wild-type HCMV infection induced only 11 of these genes at 6 h after infection (Fig. 3B). The extent to which IFN-responsive RNAs were induced by Δpp65 relative to wild-type HCMV varied significantly from gene to gene, ranging from 1.2- to >90-fold. This is consistent with the Northern blot assay displayed in Fig. 2B. The selective nature of pp65 action suggests that it targets a component of the IFN response pathway that is important for the regulation of a subset of responsive genes, but not for the pathway as a whole.
Table 1. IFN response genes.
| Average fold up-regulation
|
|||
|---|---|---|---|
| Gene | HCMV | Δpp65 | Ratio |
| IFN-γ-inducible early response gene | 6.5 ± 2.2 | 589.6 ± 57.7 | 90.8 |
| IFN-γ treatment inducible mRNA | 0.7 ± 0.1 | 26 ± 0 | 37.2 |
| IFN-β-2 | 3.2 ± 0.8 | 111.5 ± 0 | 34.9 |
| Cig5 | 35.7 ± 5.3 | 461.8 ± 22.2 | 13 |
| Monocyte chemotactic protein 2 | 1.5 ± 0.3 | 14 ± 0 | 9.4 |
| Guanylate binding protein isoform I (GBP-2) | 3.2 ± 0.5 | 28.9 ± 1.5 | 9.1 |
| GTP cyclohydrolase I | 6.8 ± 3.5 | 60.3 ± 11.8 | 8.9 |
| IFN-γ-inducible indoleamine 2,3-dioxygenase | 2.6 ± 0.7 | 22.7 ± 2.3 | 8.8 |
| 1.6-kb mRNA for 2-5A synthetase induced by IFN | 14.5 ± 2.2 | 102 ± 24.8 | 7.1 |
| 2-5 oligoadenylate synthetase 59 kDa | 20.7 ± 5.1 | 115.5 ± 5.7 | 5.6 |
| Human p78 protein | 9.4 ± 4.0 | 48.6 ± 0 | 5.2 |
| Hepatitis C-associated microtubular aggregate protein p44 | 2.7 ± 0.6 | 11.8 ± 0.6 | 4.4 |
| IFN-inducible γ2 protein | 1.9 ± 0.1 | 8.3 ± 0.5 | 4.4 |
| MxB | 2.4 ± 0.2 | 9.6 ± 0.5 | 4 |
| IFN regulatory factor 1 | 1.6 ± 0.3 | 5.7 ± 0.6 | 3.6 |
| Oligo(A) synthetase E gene | 8.6 ± 0 | 28.9 ± 1.5 | 3.4 |
| Homo sapiens cDNA, 5 end/clone = IMAGE-446622 | 11.6 ± 3.4 | 36.8 ± 0 | 3.2 |
| RING4 | 1.9 ± 0.1 | 5.5 ± 0.3 | 2.9 |
| X87344: H. sapiens DMA, DMB, HLA-Z1, IPP2, LMP2, TAP1 | 1.2 ± 0.1 | 3.4 ± 0.2 | 2.9 |
| H. sapiens mRNA; cDNA DKFZp586E0518 | 1.4 ± 0.2 | 3.9 ± 0.2 | 2.8 |
| H. sapiens mRNA expressed in osteoblast | 1.8 ± 0.8 | 4.8 ± 0.3 | 2.7 |
| 69-kDa 25 oligoadenylate synthetase | 1.4 ± 0.1 | 3.2 ± 0.5 | 2.3 |
| IFN-γ-inducible protein (IP-30) | 1.5 ± 0 | 3.4 ± 0.2 | 2.3 |
| Cig49 | 48.7 ± 4.8 | 104 ± 0 | 2.2 |
| Human nuclear phosphoprotein mRNA | 3.6 ± 1.6 | 7.7 ± 2.3 | 2.2 |
| 71-kDa 25 oligoadenylate synthetase | 2.5 ± 0.6 | 5.2 ± 0.8 | 2.1 |
| ISG-54K | 148.5 ± 29 | 304.7 ± 15 | 2.1 |
| DNA sequence from clone 494G10 on chromosome 22 | 2 ± 0.8 | 3.3 ± 0.4 | 1.7 |
| IFN-inducible protein (AIM2) | 2.8 ± 1.1 | 4.2 ± 0.7 | 1.5 |
| ICB-1 | 10.3 ± 0.5 | 12.3 ± 2.4 | 1.2 |
pp65 suppresses expression of IFN-response genes in HCMV-infected cells. DNA array analysis was carried out on duplicate samples of RNA prepared from fibroblasts 6 h after infection with wild-type HCMV or Δpp65. The fold changes for the genes shown were averaged, and the standard deviation was calculated. The “Ratio” column is the average fold up-regulation in response to Δpp65 divided by the average fold up-regulation in response to wild-type HCMV.
Table 2. Proinflammatory chemokines.
| Average fold up-regulation
|
|||
|---|---|---|---|
| Gene | HCMV | Δpp65 | Ratio |
| Chemokine exodus-1 | 0.3 ± 0.9 | 23 ± 9.8 | 76.7 |
| RANTES | 1.5 ± 0.5 | 76.2 ± 3.8 | 50.8 |
| GRO-β | 2.3 ± 1.1 | 48 ± 22.7 | 20.9 |
| Chemokine α 3 | 0.9 ± 0.3 | 11 ± 1.7 | 12.3 |
| Monocyte chemotactic protein 2 | 1.5 ± 0.3 | 14 ± 0 | 9.4 |
| Monocyte chemotactic protein 3 | 0.9 ± 0.5 | 6.8 ± 0.4 | 7.6 |
| GRO-γ | 0.8 ± 0.9 | 4.2 ± 0.7 | 5.3 |
| IL-1-β | 2.9 ± 0.3 | 8.2 ± 2.4 | 2.9 |
| Mig | 3.7 ± 0.4 | 10.7 ± 2.1 | 2.9 |
pp65 suppresses expression of proinflammatory chemokines in HCMV infected cells. DNA array analysis was carried out on duplicate samples of RNA prepared from fibroblasts at 6 h after infection with wild-type HCMV or Δpp65. The fold changes for the genes shown were averaged and the standard deviation calculated. The “Ratio” column is the average fold up-regulation in response to Δpp65 divided by the average fold up-regulation in response to wild-type HCMV.
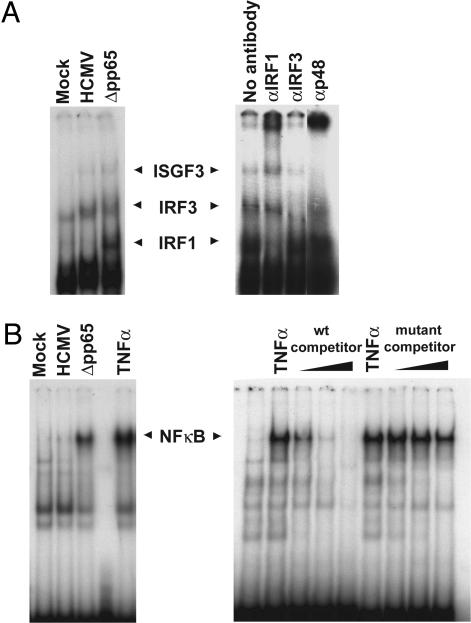
pp65 Inhibits IRF1 and NF-κB Activity in HCMV-Infected Cells. As noted above, IFN-α and β responsive genes can be induced by the binding of ISGF3 (a complex of Stat1, Stat2, and p48) to ISRE sequences in their transcriptional regulatory regions. During infection with viruses, a number of other factors, such as IRF1 and IRF3, also can bind to ISREs and regulate transcription (17, 18). Furthermore, NF-κB participates in the activation of many IFN response genes and proinflammatory chemokine genes (19, 20). To determine whether pp65 was functioning by targeting any of these factors, we analyzed their activities at 6 h after infection with Δpp65 as compared with wild-type virus. DNA-binding activities were measured by EMSA, protein levels were assayed by Western blot, and protein locations were determined by immunofluorescence.
Three distinct complexes were produced when nuclear extracts from infected cells were incubated with a 32P-labeled ISRE (Fig. 4A Left). Their relative sizes, as well as supershift assays performed with antibodies that recognize factors known to bind to ISREs, identified the complexes as IRF1, IRF3, and ISGF3 (Fig. 4A Right).
Fig. 4.
The activation of IRF1 and NF-κB is inhibited by pp65. Fibroblasts were infected with wild-type HCMV or Δpp65, and nuclear protein extracts were prepared 6 h later. The extracts were incubated with 32P-labeled oligonucleotides containing an ISRE motif (A Left) or an NF-κB-binding site (B Left) and resolved on a nondenaturing polyacrylamide gel. To confirm the identity of the complexes binding the ISRE, supershift assays were performed on a Δpp65-infected nuclear extract by using antibodies to the indicated polypeptide constituents of ISRE-binding activities (A Right). To confirm the identity of the complex binding the NF-κB probe, a competition assay was carried out (B Right). Nuclear extracts from cells treated with tumor necrosis factor α were incubated with the 32P-labeled NF-κB binding site probe plus either an unlabeled competitor DNA with the wild-type motif or a motif with a mutation that abolishes NF-κB binding.
Infection with the Δpp65 mutant consistently caused a slightly stronger (≈2-fold) recruitment of ISGF3 to the ISRE than did infection with wild-type HCMV (Fig. 4A Left), suggesting that pp65 might exert a mild inhibitory effect on the DNA-binding activity of this transcription factor. A Western blot assay showed that the levels of Stat1 and Stat2 phoshorylation were equivalent after infection with mutant or wild-type virus (Fig. 5), and an immunofluorescence assay showed that the two Stat proteins were relocated to the nucleus by 6 h after infection with either virus (Fig. 6). We conclude that pp65 does not inhibit antiviral gene expression by preventing Stat1/2 phosphorylation or nuclear translocation, and it seems unlikely that the modest difference in ISGF3 binding could explain the dramatic increases in expression levels for some IFN response genes seen in Δpp65 as compared with wild-type infections (Tables 1 and 2).
Fig. 5.
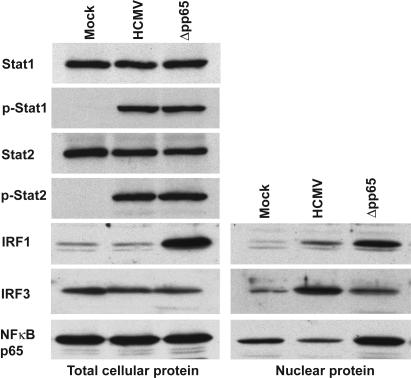
Western blot assays of factors known to regulate IFN-responsive genes. Fibroblasts were infected with wild-type HCMV or Δpp65. At 6 h after infection, cells were lysed and extracts of total cell protein (Left) and nuclear protein (Right) were prepared. The protein samples were then analyzed by Western blot assays using antibodies specific for the indicated proteins.
Fig. 6.
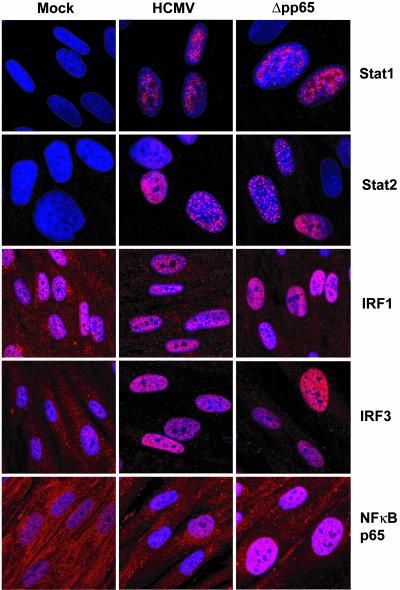
Immunofluorescent localization of factors known to regulate IFN-responsive genes. Fibroblasts grown on coverslips were infected with wild-type HCMV or Δpp65. At 6 h after infection, the cells were fixed, permeabilized, and stained with antibodies to the proteins indicated (red). Nuclei were stained with DAPI (blue).
Recruitment of IRF3 in nuclear extracts to the ISRE was not greater after infection with Δpp65 than with wild-type virus (Fig. 4A), indicating that pp65 does not inhibit this factor's DNA-binding activity. Furthermore, Western blot (Fig. 5) and immunofluorescent (Fig. 6) assays on infected cells showed that a similar amount of IRF3 relocated to the nuclear compartment after infection with HCMV or Δpp65.
IRF1 binding to the IRSE was substantially enhanced in nuclear extracts prepared after infection with the Δpp65 mutant as compared with wild-type virus (Fig. 4A). Furthermore, the amount of IRF1 protein was markedly induced after infection with Δpp65 as compared with wild-type virus, and, correspondingly, IRF1 accumulation in nuclear extracts was greater for Δpp65 (Fig. 5). Immunolocalization showed accumulation of IRF1 in the nucleus for both viruses (Fig. 6), but the difference in protein levels for the two viruses that was evident in the Western blot assay was not as apparent in their fluorescent intensities.
Finally, we examined NF-κB activity in Δpp65 versus wild-type virus-infected cells. Previous studies have shown that HCMV infection rapidly activates NF-κB (21, 22). Consistent with these reports, at 6 h after infection we observed the formation of a complex that shifted a 32P-labeled oligonucleotide containing a NF-κB binding site probe (Fig. 4B Left). Tumor necrosis factor α, a known activator of NF-κB, induced the formation of a complex that exhibited identical migration, and the identity of the complex was further confirmed by performing a competition assay with unlabeled oligonucleotides that either corresponded to the labeled probe or contain a mutation that abolishes NF-κB binding (Fig. 4B Right). Strikingly, the level of NF-κB DNA-binding activity was much greater in nuclear extracts from cells infected with Δpp65 as compared with its wild-type parent (Fig. 4B Left), indicating that pp65 has a strong inhibitory effect on the activation of NF-κB in infected cells. A Western blot assay demonstrated that the total level of the NF-κB p65 subunit was not altered by infection with either virus, but the mutant caused a more substantial relocation of NF-κB p65 to the nuclear compartment than did the wild-type virus (Fig. 5). An immunolocalization assay confirmed this result (Fig. 6).
Discussion
The importance of the IFN system in controlling viral infection is attested to by the many diverse mechanisms that viruses have evolved to antagonize it. For example, the adenovirus E1A protein inhibits IFN signaling (23) and the adenovirus virus-associated RNAs block activity of the IFN-inducible protein kinase R (PKR) (24), myxoma virus expresses a soluble decoy IFN-γ receptor (25), and the vaccinia virus E3L protein blocks the activity of PKR as well as the unidentified kinase that phosphorylates IRF3 (26). Among the herpes viruses, the Epstein–Barr virus EBNA-2 protein blocks IFN signal transduction (27) and the EBER RNAs inhibit activation of PKR (28). Two herpes simplex virus type 1 proteins, ICP0 and γ34.5, target distinct aspects of the IFN pathway. ICP0 counteracts a block to viral transcription, whereas γ34.5 targets a block to translation (29, 30).
We have previously shown that HCMV infection induces the 58-kDa DNAJ-C3 protein, a natural inhibitor of PKR, presumably to avoid the antiviral effects of this kinase (10). We also speculated that in the first few hours of infection, HCMV synthesizes a protein that suppresses the induction of IFN-responsive genes, because the block is partially relieved when cells are infected with UV-inactivated virus that is unable to express proteins from its genome (10). Here we show that, in addition to the block mediated by a newly expressed viral protein, HCMV delivers a virion protein to cells that antagonizes the induction of a subset of IFN-responsive genes (Fig. 1).
UL83-coded pp65 interferes with IFN signaling in cells where it is expressed (Fig. 2A) and it is required to suppress the expression of IFN-responsive genes in HCMV-infected cells (Fig. 2B). DNA array analysis showed that, in the absence of pp65, HCMV infection more strongly induced the accumulation of many RNAs encoded by IFN responsive genes (Table 1) and proinflammatory chemokine genes (Table 2). These data argue that pp65, which rapidly moves to the nucleus after it is delivered to infected cells as a virion component (7, 31), suppresses antiviral cellular gene expression at the start of infection.
It is notable that the effect of deleting pp65 from the viral genome, with respect to changes in the expression of IFN-response genes (Tables 1 and 2), is similar to that resulting from UV-inactivation of the viral particle (10). However, pp65 is not likely the target of UV-inactivation. First, there are several differences in the profiles of IFN-responsive genes induced by the two conditions. For example, IFN-inducible peptide 6–16 and toll-like receptor 3 are induced by UV-treated virus but not pp65-deficient virus, whereas IFN-γ treatment inducible protein is induced by pp65-deficient virus but not by UV-inactivated virus. Second, there are many differences in the effects of the UV-inactivated virus as compared with the pp65-deficient virus on the accumulation of non-IFN-responsive RNAs. Third, pp65 is not significantly expressed from the viral genome at 6 h after infection when its effect has been observed. It is conceivable that pp65 protein function is damaged or altered by UV treatment of virions. However, we favor the interpretation that a second, as yet unidentified, viral protein that is expressed de novo from the viral genome after infection also inhibits antiviral gene expression. Expression of this protein would be blocked by UV treatment. Presumably, the de novo-expressed viral protein and pp65 normally work in concert to antagonize the cellular antiviral response.
In light of the role that pp65 plays in suppressing activation of the IFN pathway, it is curious that Δpp65 exhibits only a small growth defect in cultured fibroblasts (11). It is possible that HCMV expresses other proteins that target downstream effects of the IFN-induced proteins, thereby allowing it to replicate efficiently in the presence of an activated IFN-response pathway. Alternatively, pp65 suppression of IFN signaling may play a more significant role in vivo, or in a different cell type. Consistent with an in vivo role, deletion of the murine cytomegalovirus homologue, M83, leads to attenuated replication of the mutant virus in infected animals (32).
Because NF-κB DNA-binding activity was greatly enhanced after infection with Δpp65 relative to infection with wild-type virus (Fig. 4B), we propose that the ability of pp65 to inhibit antiviral gene expression is largely mediated by blocking the activation of NF-κB. IRF1 activity also was enhanced after infection with the pp65-deficient virus, but, because expression of IRF1 is itself IFN responsive, it is likely that its induction is a secondary consequence of the block rather than a primary target.
Several reports have shown that NF-κB is activated by infection with wild-type HCMV, and NF-κB has been proposed to play an accessory role in activation of viral gene expression (16, 17, 33). Indeed, the HCMV major immediate–early promoter contains multiple NF-κB binding sites (34). Furthermore, the induction of NFκB has been proposed to block apoptosis in herpes simplex type 1-infected cells (35), and it could conceivably do the same in HCMV-infected cells. Why would HCMV antagonize a cellular factor if it promotes viral replication? Possibly, the virus induces an intermediate level of NF-κB activity, a level that is sufficient to promote viral replication, but does not fully induce antiviral genes.
Our observation that only a subset of IFN-responsive genes are induced after mutation of pp65 (Table 1) is consistent with a model in which pp65 acts by targeting NF-κB. Many IFN-responsive genes are regulated by a combination of several different factors including NF-κB (36). It is likely that the genes showing the greatest difference in expression level between wild-type HCMV and Δpp65 are the genes whose expression is most dependent on NF-κB. The mechanism by which pp65 inhibits the full activation of NF-κB is unclear. It could act indirectly to prevent its release from IκBinthe cytoplasm, or it could promote the degradation of NF-κB or its removal from the nucleus.
Acknowledgments
We thank J. Nelson (Oregon Health Sciences University, Portland) for the kind gift of Δpp65. This work was supported by National Institutes of Health Grants CA82396 and CA87661.
Abbreviations: HCMV, human cytomegalovirus; Stat, signal transducer and activator of transcription; ISRE, IFN-stimulated response element; DAPI, 4′,6-diamidino-2-phenylindole; ISGF3, IFN-stimulated gene factor 3.
References
- 1.Mendelson, M., Monard, S., Sissons, P. & Sinclair, J. (1996) J. Gen. Virol. 77, 3099–3102. [DOI] [PubMed] [Google Scholar]
- 2.Goodrum, F. D., Jordan, C. T., High, K. & Shenk, T. (2002) Proc. Natl. Acad. Sci. USA 99, 16255–16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarvis, M. A. & Nelson, J. A. (2002) Curr. Opin. Microbiol. 5, 403–407. [DOI] [PubMed] [Google Scholar]
- 4.Ahn, K., Angula, A., Chazal, P., Yang, Y. & Fruh, K. (1996) Proc. Natl. Acad. Sci. USA 93, 10990–10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutermann, A., Bubeck, A., Wagner, M., Reusch, U., Menard, C. & Koszinowski, U. H. (2002) Curr. Top. Microbiol. Immunol. 269, 1–22. [DOI] [PubMed] [Google Scholar]
- 6.Kotenko, S. V., Saccani, S., Izotova, L. S., Mirochnitchenko, O. V. & Pestka, S. (2000) Proc. Natl. Acad. Sci. USA 97, 1695–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu, H., Cong, J. P. & Shenk, T. (1997) Proc. Natl. Acad. Sci. USA 94, 13985–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu, H., Cong, J. P., Mamtora, G., Gingeras, T. & Shenk, T. (1998) Proc. Natl. Acad. Sci. USA 95, 14470–14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle, K. A., Pietropaolo, R. L. & Compton, T. (1999) Mol. Cell. Biol. 19, 3607–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browne, E. P., Wing, B., Coleman, D. & Shenk, T. (2001) J. Virol. 75, 12319–12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmolke, S., Kern, H. F., Drescher, P., Jahn, G. & Plachter, B. (1995) J. Virol. 69, 5959–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anonymous (2000) Affymetrix Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA).
- 13.Sun, Z., Arendt, C. W., Ellmeier, W., Schaeffer, E. M., Sunshine, M. J., Gandhi, L., Annes, J., Petrzilka, D., Kupfer, A., Schwartzberg, P. L., et al. (2000) Nature 404, 402–407. [DOI] [PubMed] [Google Scholar]
- 14.Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H. & Schreiber, R. D. (1998) Annu. Rev. Biochem. 67, 227–264. [DOI] [PubMed] [Google Scholar]
- 15.Levy, D. E. & Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662. [DOI] [PubMed] [Google Scholar]
- 16.Miller, D. M., Zhang, Y., Rahill, B. M., Waldman, W. J. & Sedmak, D. D. (1999) J. Immunol. 162, 6107–6113. [PubMed] [Google Scholar]
- 17.Parrington, J., Rogers, N. C., Gewert, D. R., Pine, R., Veals, S. A., Levy, D. E., Stark, G. R. & Kerr, I. M. (1993) Eur. J. Biochem. 214, 617–626. [DOI] [PubMed] [Google Scholar]
- 18.Grandvaux, N., Servant, M. J., tenOever, B., Sen, G. C., Balachandran, S., Barber, G. N., Lin, R. & Hiscott, J. (2002) J. Virol. 76, 5532–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genin, P., Algarte, M., Roof, P., Lin, R. & Hiscott, J. (2000) J. Immunol. 164, 5352–5361. [DOI] [PubMed] [Google Scholar]
- 20.Kim, T. K. & Maniatis, T. (1997) Mol. Cell 1, 119–129. [DOI] [PubMed] [Google Scholar]
- 21.Yurochko, A. D., Kowalik, T. F., Huong, S. M. & Huang, E. S. (1995) J. Virol. 69, 5391–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yurochko, A. D., Hwang, E. S., Rasmussen, L., Keay, S., Pereira, L. & Huang, E. S. (1997) J. Virol. 71, 5051–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reich, N., Pine, R., Levy, D. & Darnell, J. E., Jr. (1988) J. Virol. 62, 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathews, M. B. & Shenk, T. (1991) J. Virol. 65, 5657–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Upton, C., Mossman, K. & McFadden, G. (1992) Science 258, 1369–1372. [DOI] [PubMed] [Google Scholar]
- 26.Smith, E. J., Marie, I., Prakash, A., Garcia-Sastre, A. & Levy, D. E. (2001) J. Biol. Chem. 276, 8951–8957. [DOI] [PubMed] [Google Scholar]
- 27.Kanda, K., Decker, T., Aman, P., Wahlstrom, M., von Gabain, A. & Kallin, B. (1992) Mol. Cell. Biol. 12, 4930–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke, P. A., Schwemmle, M., Schickinger, J., Hilse, K. & Clemens, M. J. (1991) Nucleic Acids Res. 19, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou, J. & Roizman, B. (1992) Proc. Natl. Acad. Sci. USA 89, 3266–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mossman, K. L. & Smiley, J. R. (2002) J. Virol. 76, 1995–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldick, C. J., Jr. & Shenk, T. (1996) J. Virol. 70, 6097–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morello, C. S., Cranmer, L. D. & Spector, D. H. (1999) J. Virol. 73, 7678–7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowalik, T. F., Wing, B., Haskill, J. S., Azizkhan, J. C., Baldwin, A. S., Jr., & Huang, E. S. (1993). Proc. Natl. Acad. Sci. USA 90, 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan, Y. J., Tseng, W. P. & Hayward, G. S. (1996) J. Virol. 70, 5312–5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodkin, M. L., Ting, A. T. & Blaho, J. A. (2003) J. Virol. 77, 7261–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doyle, S. E., Vaidya, S. A., O'Connell, R., Dadgostar, H., Dempsey, P. W., Wu, T.-T., Rao, G., Sun, R., Haberland, M. E., Modlin, R. L., et al. (2002) Immunity 17, 251–263. [DOI] [PubMed] [Google Scholar]