Abstract
Here we exploit the extensive cell lineage information and streamlined genome of the ascidian, Ciona intestinalis, to investigate heart development in a basal chordate. Several cardiac genes were analyzed, including the sole Ciona ortholog of the Drosophila tinman gene, and tissue-specific enhancers were isolated for some of the genes. Conserved sequence motifs within these enhancers facilitated the isolation of a heart enhancer for the Ciona Hand-like gene. Altogether, these studies provide a regulatory framework for the differentiation of the cardiac mesoderm, beginning at the 110-cell stage, and extending through the fusion of cardiac progenitors during tail elongation. The cardiac lineage shares a common origin with the germ line, and zygotic transcription is first detected in the heart progenitors only after its separation from the germ line at the 64-cell stage. We propose that germ-line determinants influence the specification of the cardiac mesoderm, both by inhibiting inductive signals required for the development of noncardiac mesoderm lineages, and by providing a localized source of Wnt-5 and other signals required for heart development. We discuss the possibility that the germ line also influences the specification of the vertebrate heart.
Recent studies have identified several signaling pathways and regulatory factors governing heart development in vertebrates (1–4). Bone morphogenetic protein (BMP), Wnt-11, and fibroblast growth factor (FGF) signaling establish localized expression of key cardiac transcription factors, such as Gata-4/5/6, Nkx-2.5, and dHand. However, understanding the molecular details of these interactions has been complicated by extensive gene duplication events in vertebrates. For example, Nkx-2.5 is a member of a gene family that includes Nkx-2.3, -2.6, -2.7, and -2.8. To circumvent this genetic redundancy, we have launched an analysis of heart development in the basal chordate, Ciona intestinalis.
In ascidians such as Ciona, the heart consists of a single layered myoepithelial U-shaped tube surrounded by a single-layered pericardial coelom (5, 6). Although rudimentary in structure, tunicate heart morphogenesis is similar to the initial stages of vertebrate heart formation (5, 7). Just before gastrulation (the 110-cell stage), the heart lineage is derived from a pair of cells near the vegetal pole (the B7.5 blastomeres). Soon after neurulation, the heart precursors consist of a set of four large bilaterally paired cells, termed the trunk ventral cells (TVCs) (8). The TVCs migrate ventrally and fuse along the midline of the larva (see below). After metamorphosis, a subset of these cells form the heart tube.
Here we present, to our knowledge, the first visualization of heart specification in Ciona, by using a combination of in situ hybridization assays and electroporation-based transgenesis methods. The recent sequencing of the C. intestinalis genome has allowed us to identify the orthologs of genes expressed in the early vertebrate heart field, including Nkx-2.5, d/e-Hand, Troponin I, and Troponin T. We have characterized the expression of these genes and identified the enhancers that direct their expression. The detailed visualization of the Ciona cardiac lineage suggests that germ-line determinants influence the initial specification of the cardiac mesoderm.
Materials and Methods
Ascidians. Ciona adults were collected from Half Moon Bay and Oyster Point (CA), and imported from various sources, including Japan (generously provided by N. Satoh, Kyoto University, Kyoto), the Station Biologique de Roscoff (Roscoff, France), and the Marine Biological Laboratory (Woods Hole, MA). No significant discrepancies in the expression of transgenes were observed among animals obtained from these different sources. Rearing, fertilization, dechorionation, in situ hybridization, electroporation, and lacZ staining were conducted as described in Corbo et al. (9).
Electroporation Constructs. LacZ fusion genes were prepared by isolating genomic DNA fragments by means of PCR amplification, and then cloning the fragments into the pCES vector, which contains the Ci-fkh promoter region and lacZ reporter gene (10). Genomic DNA was isolated from pooled sperm of three to four adults by using the PureGene DNA isolation kit (Gentra Systems). Final insert products were amplified with the following oligonucleotide primer pairs (numbers indicate base pair distance 5′ of the EST-predicted transcript): TropIF986-TCAGCAGGACTTTACTTTTCGTGG, TropIB76-GGTTGCATTCAGTTACTG, NPPF1073-CGGATCTCCGATTAATG, NPPB36-GTCGCAATGCATGAG, TropTF657-GCAACCAGATGC, TropTB49-GGTCACTGGACCATGG, 29h10F910-CCTACTGTATTCCATGC, 29h10B376-TTCAACCCCAAACGCATCC, HndxF2955-CGTTCTTGAAACTGGAGACATCCTG, and HndxB58-GCCACGTGAAACTA. The TropI, Npp, Hndx, and 29h10 primers included XbaI sites on the forward primers and BamHI sites on the reverse primers, allowing the Npp and Hndx fragments to be cloned directly into the XbaI/BamHI sites of the pCES vector. The TropI insert was subcloned, employing the 3′ BamHI site and a 5′ internal SphI site (827 bp from the transcription start site). The 29h10 and TropT DNA fragments were first ligated onto the 3′ T overhangs of the pGEM-T Easy vector (Promega). They were then subcloned into pCES by using restriction sites in the polylinker sequence in the pGEM vector. A PstI site was used for the 29h10 enhancer (along with the PCR-generated 3′ BamHI site), and a 5′ PstI site and 3′ NcoI site were used for the TropT enhancer.
Confocal Microscopy. Larvae hybridized to the Ci-SercaA antisense digoxigenin (DIG) probe were processed according to standard procedures (9). After incubation with the anti-DIG antibody, they were rinsed four times in PBT (126 nM NaCl/3 mM NaH2 PO4·H2O/7 mM Na2HPO4/0.01% Tween 20) than rinsed once with a Fast red staining buffer (1 M Tris, pH 8.2/1% Tween 20), stained with Fast red (Roche) in the same buffer for 1–2 h, rinsed with PBT and mounted in PBT:Glycerol. Confocal images were obtained on a Zeiss 510 laser scanning confocal microscope (courtesy of the CNR biological imaging facility at the University of California, Berkeley). Images consisting of projected Z series were processed by using the bitplane imaris 3.3 software package.
Comparative Analysis of Enhancers. The codegrok bioinformatics package was used to identify shared sequence motifs that were overrepresented within different heart enhancers (11). Aspects of the software are similar to publicly available search algorithms, such as moby dick (http://genome.ucsf.edu:8080/mobydick).
Results
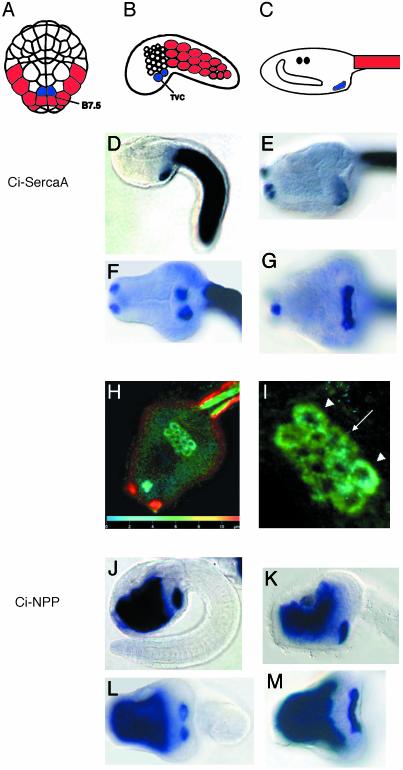
At the 110-cell stage, the progenitors of the adult heart consist of a single pair of cells, the B7.5 blastomeres (Fig. 1A). At the onset of tailbud formation, a subset of B7.5 daughter cells form the four bilaterally paired TVCs (Fig. 1B). These cells divide and fuse to form a continuous band of cells (Fig. 1 G and M).
Fig. 1.
Gene expression in the heart lineage. (A–C) Diagrams of Ciona embryos and larvae. Tail muscle myoblasts are red, and heart lineage cells (TVCs) are blue. The anterior is up (A) to the left (B and C). (D–M) Ciona larvae stained with antisense RNA probes for Ci-SercaA (ci0100137099, clone ID no. GC29o11) (D–I) and Ci-NPP (ci0100144439, clone ID no. GC29e18) (J–M). The top rows are side views of tailbud embryos (D and J) and larval heads (E and K). The bottom rows are ventral views of tailbud embryos (F and L) and larval heads (G and M). (H) Ventral view of larva hybridized to Ci-SercaA and stained with Fast red. Projections of confocal z series false colored to represent depth are shown (scale on the z axis is indicated on bar at the bottom of the image). (I) Closeup (from H) of emerging heart field. Arrowheads point to four large outer cells, and the arrow points to inner cluster of eight smaller cells. To obtain sequence information, use the gene numbers provided above to search within the annotation section (http://genome.jgi-psf.org/ciona4/ciona4.home.html), or use the clone ID no. to search the EST site (http://ghost.zool.kyoto-u.ac.jp/indexr1.html).
A variety of gene markers permit the detailed visualization of the TVCs through in situ hybridization; two are shown here (Fig. 1 D–M). ESTs obtained from the Ciona intestinalis Gene Collection Release 1 (12) were used to prepare DIG-labeled antisense RNA probes for in situ hybridization [see Corbo et al. (9)]. One of the markers, Ci-SercaA, encodes a calcium-ATPase that is expressed in the TVCs, papillae, and tail muscles (Fig. 1 D–I). The other marker gene, Ci-NPP, encodes a phosphodiesterase that is expressed in the TVCs and endoderm (Fig. 1 J–M). During early phases of tailbud formation, the TVCs consist of two bilateral clusters of cells (Fig. 1 F and L), but by the completion of tail elongation, these clusters fuse to form a single band of cells (Fig. 1 G, H, and M).
Fluorescent visualization of Ci-SercaA expression permitted a three-dimensional reconstruction of the larval heart field (Fig. 1 H and I). Confocal visualization involved embryos hybridized with a DIG-labeled antisense RNA probe, which were then stained with Fast red (see Materials and Methods). The larval cardiac cells consisted of a flat sheet, showing no evidence of heart tube formation. Additionally, there are two distinct cell populations at this point, a central group of eight small cells, and an outer group of four large cells. This discrepancy in cell size/number can be easily explained through asymmetrical mitosis of the central vs. peripheral founder TVCs.
A survey of the Ciona genome reveals a single ortholog of the vertebrate Nkx-2.5 gene (Ci-Nkx). Because of the importance of this gene in the development of cardiac tissues in both vertebrates and Drosophila (tinman), we have examined the expression profile of Ci-Nkx in detail (Fig. 2).
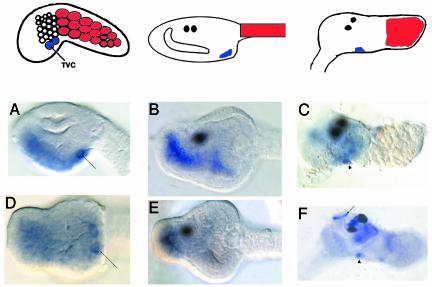
Fig. 2.
Ci-Nkx expression. The top row displays diagrams of representative stages as in Fig. 1. (Right Upper) The diagram portrays a juvenile ≈36 h after metamorphosis, with the resorbed tail muscle in red. The anterior is to the left in all diagrams and pictures. (A–E) Ciona larvae stained with antisense RNA probes against Ci-Nkx (ci0100140552, clone ID no.GC29k02). (A–C) Side views of a tailbud head (A), larval head (B), and a juvenile (C) ≈36 h after metamorphosis. Note staining of the differentiating heart (arrowhead). Contractile activity in the heart can first be detected ≈60 h after metamorphosis. Also shown are ventral views of a tailbud head (D) and a larval head (E). Note staining of the TVCs in A and D (arrow). We have also included a side view of an ≈36-h-old juvenile stained with Ci-TnI (F). Note staining of the siphon muscles (arrow), along with the heart (arrowhead).
Ci-Nkx expression is first detected in the TVCs after neurulation. By the tailbud stage, staining is detected in the TVCs, along with the ventral endoderm and ventral trunk epidermis (Fig. 2 A and B). This staining of the ventral tissues is already apparent during neurulation (data not shown), but TVC expression is not detectable until the tailbud stage. Ventral views of elongating tailbud embryos clearly reveal the expression of the Ci-Nkx gene in the TVCs (Fig. 2D). This staining is lost by the completion of elongation, but is maintained in the anterior endoderm (Fig. 2 B and E). In contrast, the expression of other cardiac marker genes, such as Ci-SercaA, persists within the heart field during larval development and metamorphosis (e.g., Fig. 1G). While lost in swimming tadpoles, Ci-Nkx expression reappears in the definitive heart after the onset of metamorphosis (Fig. 2C, arrowhead). Expression in the TVCs, anterior endoderm, and ventral ectoderm is comparable to the expression profile of Nkx-2.5 in vertebrates (13, 14). In contrast, persistent tinman expression is restricted to the mesoderm in Drosophila embryos (15).
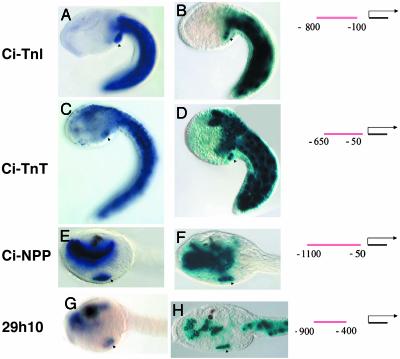
To outline the genetic network underlying Ciona heart development, we have identified tissue-specific enhancers for a number of genes that are expressed in the TVCs. Several examples are presented in Fig. 3. The Ciona homologs of the vertebrate Troponin I (Ci-TnI) and Troponin T (Ci-TnT) genes are expressed in the tail muscles and TVCs (Fig. 3 A and C). The Ci-TnT gene is also expressed in the CNS, including the cerebral vesicle (Fig. 3C). Short genomic DNA fragments from the 5′ flanking regions of these genes direct authentic patterns of lacZ expression in electroporated embryos (Fig. 3 B and D). For example, an ≈700-bp fragment from Ci-TnI was cloned into a reporter gene containing the minimal Ci-Fkh promoter region and lacZ. The fusion gene was electroporated into one-cell embryos and then visualized by X-Gal staining. Expression is restricted to the tail muscles and TVCs (Fig. 3B), as seen for the endogenous gene (Fig. 3A). An ≈600-bp genomic DNA fragment from Ci-TnT is sufficient to direct lacZ expression in the TVCs, tail muscles, and CNS (Fig. 3D; compare with C).
Fig. 3.
Enhancers for Ciona heart genes. (A, C, E, and G) In situ staining for four probes, as indicated to the left, Ci-TnI (ci0100137955, clone ID no. GC29b15), Ci-TnT (ci0100146672, clone ID no. GC26c17), Ci-NPP, and 29h10 (ci0100135212, named after clone ID no. GC29h10). (B, D, F, and H) Corresponding lacZ reporter assays for regulatory DNA located upstream of the relevant genes. Diagrams to the right indicate the position of the tested fragment relative to the predicted transcriptional start site of the relevant gene. Note staining of the TVC progeny in all images (arrowheads). Note staining in the tail muscle in A–D, staining in the endoderm in E–H, and staining of the CNS in C–D. Also note that the 29h10 enhancer drives expression in the tail muscle (H), although this is not seen in the endogenous expression pattern (G). (A–D) Side view of tailbud embryos. (E–H) Side view of larval heads. The anterior is to the left in all images.
Tissue-specific enhancers were also identified for two genes that are expressed in both the endoderm and TVCs; Ci-NPP (Fig. 3E) and 29h10 (Fig. 3G), a transcript with no match to known genes (name indicates EST Collection number). An ≈1,000-bp fragment upstream of Ci-NPP directs lacZ staining in the endoderm and TVCs (Fig. 3F; compare with E); an ≈500-bp fragment upstream of 29h10 directs a similar pattern of expression (Fig. 3H; compare with G).
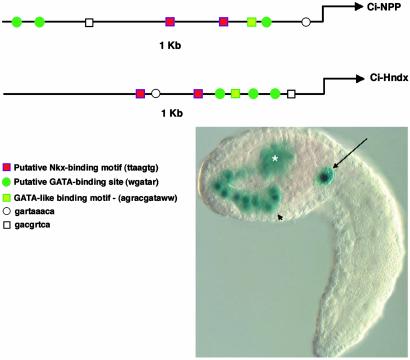
The Ciona genome contains two possible orthologs of the vertebrate Hand genes, Ci-Hnd and Ci-Hndx (Ci-Hand-like) (16). The Hand genes are crucial for vertebrate heart and limb development (17, 18). Ci-Hnd contains extensive sequence conservation with the vertebrate Hand genes, whereas Ci-Hndx is quite divergent (16). Nonetheless, Ci-Hndx is expressed in the TVCs at the tailbud stage [along with transient expression in the trunk lateral cells and endoderm at earlier stages, (19), referred to as Ci-NoTrlc]. Given the overlapping expression of Ci-NPP and Ci-Hndx in the TVCs and endoderm, bioinformatics methods were used to identify shared sequence motifs within their 5′ flanking regions. These methods led to the detection of putative Tinman- and GATA-binding sites within the defined Ci-NPP enhancer. A similar cluster of sites was identified within the first 1 kb of the Ci-Hndx 5′ flanking region (Fig. 4). A DNA fragment containing the predicted Ci-Hndx enhancer directs an authentic pattern of lacZ expression in the TVCs, trunk lateral cells, and endoderm (Fig. 4). Future studies will determine whether these sites are essential for restricted expression in the cardiac lineage.
Fig. 4.
Prediction of the Ci-Hndx enhancer. (Upper) Alignment of proximal 1 kb of upstream sequence for Ci-NPP and Ci-Hndx (ci0100140298, clone ID no. GC31k05) showing conservation of clustered motifs. (Left Lower) Sequence and tentative designation of motifs is shown. (Right Lower) A typical staining pattern for the 5′ region upstream of Ci-Hndx attached to Fkh-lacZ. Note staining in the endoderm (arrowhead), trunk lateral cells (asterisk), and TVCs (arrow).
We have also characterized the expression of Ci-Wnt5 (the sole Ciona ortholog of the Wnt-5a class genes, which includes Wnt-11) (20). As in Halocynthia (21), Ciona maternal Wnt-5 mRNA is segregated with the pole plasm (PP) (Fig. 5C). Vertebrate Wnt-11 is also maternally segregated to the vegetal pole, along with the germinal plasm (22).
Fig. 5.
Model of ascidian heart specification. (A and B) Diagrams of 32- and 64-cell embryos seen from the vegetal side. In these diagrams, the PP is purple and the PVC is outlined in red. Gray represents the expression patterns of Ci-Sna and Cs-ZicL. Note that when expression is limited to the nuclei (as in the B7.5 cells at the 64-cell stage), this indicates recently initiated transcription (29). (C) Antisense Ci-Wnt5 (ci0100135747, clone ID no. GC12l09) staining of a 110-cell embryo, on the vegetal side. (D) A 110-cell embryo in which descendents of the posterior vegetal quadrant have been color-coded to correspond with the cell fates listed in the diagrams to the right. (E–G) Diagrammatic representations of the posterior vegetal quadrant of the Ciona embryo. (E) At the 32-cell stage, the posteriormost blastomere (B6.3) still contains PP, repressing transcription, and thereby inhibiting a response to PVC-mediated myogenesis, as well as inductive Fgf signaling from the endoderm (yellow arrows). (F) At the 64-cell stage, the newly born B7.5 cells have now emerged from the PP and are therefore newly competent to respond to inductive signals. Possible signaling, which may occur after the 64-cell stage, is indicated by arrows, including the expression of BMP by the B7.5 cells at the 110-cell stage (red arrow) (34), FGF-8 signaling from the mesenchyme (MES) (green arrow; unpublished data), Wnt-5 signaling from the germ cell lineage (purple), or signaling from the endoderm (ENDO) (yellow). (G) By the 180-cell stage, the TVC lineage has separated from the tail muscle (MUS) lineage (35). It is unclear to what extent the TVCs are prespecified to from the heart at this point; later positional signaling may play a significant role in heart lineage specification.
Discussion
We have documented the expression of a variety of marker genes in the cardiac mesoderm of Ciona embryos. These marker genes permitted the detailed visualization of the heart lineage during development. Bilateral clusters of cardiac progenitors undergo fusion during tail elongation, and the resulting heart field contains two distinct cell types, which might arise from asymmetric mitotic divisions (Fig. 1 H and I). Asymmetric divisions may represent an early step in the distinction between internal myocardial cells and outer pericardial cells. This result would parallel the positioning of cardiac/pericardial cells in the developing Drosophila dorsal vessel (23).
Our description of Ci-Nkx expression completes a survey of tinman-related genes among the major chordate lineages (24). The expression of Ci-Nkx in paired heart primordia, the anterior endoderm, and ventral ectoderm is similar to the expression profiles observed for vertebrate Nkx2–5 genes (13, 14). In contrast, Drosophila tinman expression persists only in the mesoderm, although there is transient expression in the foregut (Berkeley Drosophila Genome Project, Berkeley, CA) (15). The sustained expression of Ci-Nkx/AmphiNk2-tin/Nkx2.5 in the anterior endoderm may relate to the development of chordate or deuterostome-specific structures, particularly the pharyngeal gill slits and endostyle.
The timing and dynamics of the Ci-Nkx expression profile is consistent with a conserved role as a key regulator of heart development. Cells within the Ciona heart field continue to divide in the larvae, but differentiate into the definitive heart only after metamorphosis. The transient down-regulation of Ci-Nkx in the larval heart field (Fig. 2 B and E) mirrors this transient arrest of heart development during the larval stage.
We have identified several regulatory DNAs that mediate gene expression in the cardiac mesoderm (Fig. 3). These DNAs share a number of short sequence motifs, including putative Tinman- and GATA-binding sites. Putative Tinman-binding sites in the Ci-Handx and Ci-NPP enhancers are consistent with a conserved regulatory role for Ci-Nkx. GATA transcription factors are essential for heart development in both Drosophila and vertebrates (2). The Ciona genome contains only two GATA factors (25). GataA is expressed in the CNS and blood-cell lineages (26), possibly regulating hematopoesis as do the vertebrate GATA 1/2/3 genes. GataB expression has not been characterized. Future testing of the function of putative GATA- and Tinman-binding motifs through site-directed mutagenesis is warranted.
Our analysis of the Ciona heart has led to an unexpected connection between germ-line determinants and the early cardiac mesoderm. The heart progenitors, TVCs, descend from a lineage positioned at the intersection of two crucial cytoplasmic determinants, the posterior vegetal cytoplasm (PVC) and the PP. Blastomeres containing PVC form either tail muscles, TVCs or mesenchyme, whereas those that inherit PP form germ cells (27, 28). At the 32-cell stage, the PVC and PP are colocalized within the B6.3 blastomeres (Fig. 5A). At the 64-cell stage, B6.3 cells divide to form anterior B7.5 daughter cells and the posterior B7.6 cells (Fig. 5B). The B7.6 cells gives rise to the germ line, whereas B7.5 cells form tail muscles and TVCs (Fig. 5 D and G). At the 32-cell stage, mesoderm-specific genes, including Ci-Sna and Cs-ZicL, are expressed in the B6.2 and B6.4 blastomeres (Fig. 5A) (29, 30), whereas expression in the progeny of B6.3 is delayed until the 64-cell stage (Fig. 5B). Therefore, there is a correlation between loss of PP and the activation of zygotic transcription. This correlation has been thoroughly documented in the ascidian Halocynthia roretzi (31).
We propose that the germ plasm contains one or more repressors that prevent B6.3 cells from responding to critical cell specification events. By the 64-cell stage, maternally loaded factors in the PVC commit B7.2 and B7.6 cells to a tail muscle cell fate, whereas FGF signals emanating from the endoderm of 32-cell stage embryos induce the B7.3 and B7.7 blastomeres to form mesenchyme (Fig. 5 E and F) (32). Germ plasm repression of B6.3 cells prevents specification by PVC or FGF. After the division of the B6.3 blastomeres, these repressors are asymmetrically localized to the B7.6 blastomeres, which form the germ-line. The B7.5 cells can now respond to subsequent cardiac specification signals (Fig. 5 F and G).
Maternal Ci-Wnt5 may have a conserved role in the specification of cardiac mesoderm. Both Ci-Wnt5 and the related Xenopus Wnt-11 gene are maternally colocalized with the germ plasm (Fig. 5C) (22). Maternal Wnt-11 is differentially translated on the dorsal side of the Xenopus embryo (33). Wnt-11-mediated signaling has recently been demonstrated as both necessary and sufficient for vertebrate cardiac mesoderm induction (3). Although it is sometimes assumed that vertebrate cardiac specification begins at gastrulation, Wnt-11 and other maternally loaded factors might prepattern the mesoderm. Such prepatterning would influence subsequent specification by inductive factors in the endoderm and organizer. Thus, it is possible that the germ plasm influences the formation of the cardiac mesoderm in both ascidians and vertebrates through conserved mechanisms.
Acknowledgments
We thank Nori Satoh, Yutaka Satou, and their colleagues for providing us a copy of the Ciona intestinalis Gene Collection Release 1, and for the remarkable database associated with this EST project (http://ghost.zool.kyoto-u.ac.jp); Didier Stainier for his review of the manuscript; and Albert Erives, Naoe Harafuji, and David Keys for their comments and insights. This work was supported by National Institutes of Health Grant 1001761 and National Science Foundation Grant 0131758 (to M.L.).
Abbreviations: TVC, trunk ventral cell; PVC, posterior vegetal cytoplasm; PP, pole plasm.
References
- 1.Andree, B., Duprez, D., Vorbusch, B., Arnold, H. H. & Brand, T. (1998) Mech. Dev. 70, 119–131. [DOI] [PubMed] [Google Scholar]
- 2.Cripps, R. M. & Olson, E. N. (2002) Dev. Biol. 246, 14–28. [DOI] [PubMed] [Google Scholar]
- 3.Pandur, P., Lasche, M., Eisenberg, L. M. & Kuhl, M. (2002) Nature 418, 636–641. [DOI] [PubMed] [Google Scholar]
- 4.Reifers, F., Walsh, E. C., Leger, S., Stainier, D. Y. & Brand, M. (2000) Development (Cambridge, U.K.) 127, 225–235. [DOI] [PubMed] [Google Scholar]
- 5.Satoh, N. (1994) Developmental Biology of Ascidians (Cambridge Univ. Press, New York).
- 6.Robb, J. S. (1965) Comparative Basic Cardiology (Grune & Stratton, New York).
- 7.de Selys-Longchamps, M. (1900) Arch. Biol. 17, 499–542. [Google Scholar]
- 8.Hirano, T. & Nishida, H. (1997) Dev. Biol. 192, 199–210. [DOI] [PubMed] [Google Scholar]
- 9.Corbo, J. C., Levine, M. & Zeller, R. W. (1997) Development (Cambridge, U.K.) 124, 589–602. [DOI] [PubMed] [Google Scholar]
- 10.Harafuji, N., Keys, D. N. & Levine, M. (2002) Proc. Natl. Acad. Sci. USA 99, 6802–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stathopoulos, A., Van Drenth, M., Erives, A., Markstein, M. & Levine, M. (2002) Cell 111, 687–701. [DOI] [PubMed] [Google Scholar]
- 12.Satoh, N., Satou, Y., Davidson, B. & Levine, M. (2003) Trends Genet. 19, 376–381. [DOI] [PubMed] [Google Scholar]
- 13.Lints, T. J., Parsons, L. M., Hartley, L., Lyons, I. & Harvey, R. P. (1993) Development (Cambridge, U.K.) 119, 419–431. [DOI] [PubMed] [Google Scholar]
- 14.Schultheiss, T. M., Xydas, S. & Lassar, A. B. (1995) Development (Cambridge, U.K.) 121, 4203–4214. [DOI] [PubMed] [Google Scholar]
- 15.Bodmer, R., Jan, L. Y. & Jan, Y. N. (1990) Development (Cambridge, U.K.) 110, 661–669. [DOI] [PubMed] [Google Scholar]
- 16.Satou, Y., Imai, K. S., Levine, M., Kohara, Y., Rokhsar, D. & Satoh, N. (2003) Dev. Genes Evol. 213, 213–221. [DOI] [PubMed] [Google Scholar]
- 17.Yelon, D., Ticho, B., Halpern, M. E., Ruvinsky, I., Ho, R. K., Silver, L. M. & Stainier, D. Y. (2000) Development (Cambridge, U.K.) 127, 2573–2582. [DOI] [PubMed] [Google Scholar]
- 18.Yamagishi, H., Yamagishi, C., Nakagawa, O., Harvey, R. P., Olson, E. N. & Srivastava, D. (2001) Dev. Biol. 239, 190–203. [DOI] [PubMed] [Google Scholar]
- 19.Imai, K. S., Satoh, N. & Satou, Y. (2003) Development (Cambridge, U.K.) 130, 4461–4472. [DOI] [PubMed] [Google Scholar]
- 20.Hino, K., Satou, Y., Yagi, K. & Satoh, N. (2003) Dev. Genes Evol. 213, 264–272. [DOI] [PubMed] [Google Scholar]
- 21.Miya, T. & Nishida, H. (2002) Dev. Genes Evol. 212, 30–37. [DOI] [PubMed] [Google Scholar]
- 22.Ku, M. & Melton, D. A. (1993) Development (Cambridge, U.K.) 119, 1161–1173. [DOI] [PubMed] [Google Scholar]
- 23.Ward, E. J. & Skeath, J. B. (2000) Development (Cambridge, U.K.) 127, 4959–4969. [DOI] [PubMed] [Google Scholar]
- 24.Holland, N. D., Venkatesh, T. V., Holland, L. Z., Jacobs, D. K. & Bodmer, R. (2003) Dev. Biol. 255, 128–137. [DOI] [PubMed] [Google Scholar]
- 25.Yamada, L., Kobayashi, K., Degnan, B., Satoh, N. & Satou, Y. (2003) Dev. Genes Evol. 213, 245–253. [DOI] [PubMed] [Google Scholar]
- 26.D'Ambrosio, P., Fanelli, A., Pischetola, M. & Spagnuolo, A. (2003) Dev. Dyn. 226, 145–148. [DOI] [PubMed] [Google Scholar]
- 27.Takamura, K., Fujimura, M. & Yamaguchi, Y. (2002) Dev. Genes Evol. 212, 11–18. [DOI] [PubMed] [Google Scholar]
- 28.Fujimura, M. & Takamura, K. (2000) Dev. Genes Evol. 210, 64–72. [DOI] [PubMed] [Google Scholar]
- 29.Erives, A., Corbo, J. C. & Levine, M. (1998) Dev. Biol. 194, 213–225. [DOI] [PubMed] [Google Scholar]
- 30.Imai, K. S., Satou, Y. & Satoh, N. (2002) Development (Cambridge, U.K.) 129, 2723–2732. [DOI] [PubMed] [Google Scholar]
- 31.Tomioka, M., Miya, T. & Nishida, H. (2002) Zool. Sci. 19, 49–55. [DOI] [PubMed] [Google Scholar]
- 32.Kim, G. J., Yamada, A. & Nishida, H. (2000) Development (Cambridge, U.K.) 127, 2853–2862. [DOI] [PubMed] [Google Scholar]
- 33.Schroeder, K. E., Condic, M. L., Eisenberg, L. M. & Yost, H. J. (1999) Dev. Biol. 214, 288–297. [DOI] [PubMed] [Google Scholar]
- 34.Miya, T., Morita, K., Suzuki, A., Ueno, N. & Satoh, N. (1997) Development (Cambridge, U.K.) 124, 5149–5159. [DOI] [PubMed] [Google Scholar]
- 35.Nishida, H. (1986) Dev. Growth Differ. 28, 191–201. [DOI] [PubMed] [Google Scholar]