Abstract
Evidence for the evolutionary maintenance of a hairpin structure possibly involved in intron processing had been found in intron 1 of the alcohol dehydrogenase gene (Adh) in diverse Drosophila species. In this study, the putative hairpin structure was evaluated systematically in Drosophila melanogaster by elimination of either side of the stem using site-directed mutagenesis. The effects of these mutations and the compensatory double mutant on intron splicing efficiency and ADH protein production were assayed in Drosophila melanogaster Schneider L2 cells and germ-line transformed adult flies. Mutations that disrupt the putative hairpin structure right upstream of the intron branch point were found to cause a significant reduction in both splicing efficiency and ADH protein production. In contrast, the compensatory double mutant that restores the putative hairpin structure was indistinguishable from the WT in both splicing efficiency and ADH level. It was also observed by mutational analysis that a more stable secondary structure (with a longer stem) in this intron decreases both splicing efficiency and ADH protein production. Implications for RNA secondary structure and intron evolution are discussed.
The neutral theory of molecular evolution holds that most of the variation observed at the molecular level is not caused by positive Darwinian selection but by random genetic drift and fixation of mutant alleles that are selectively neutral or nearly neutral (1). Mutations occurring in noncoding regions, as well as synonymous substitutions in coding regions, are regarded effectively neutral because they do not lead to amino acid changes (2). However, selection on noncoding and silent sites is also possible. Because mRNA undergoes transcription, splicing, transportation, and translation and can be regulated at many steps, it is not surprising that the information on mRNA regulation can be found not only at the DNA sequence level but also in higher-order RNA structures.
The mRNA precursors of most genes in higher eukaryotes contain introns that need to be removed by nuclear pre-mRNA splicing (3). Even though much is known about the biochemical pathway of splicing, the mechanism of accurate and efficient splice site choice remains elusive. A few consensus elements, such as the 5′ and 3′ splice sites, the branch point, and the polypyrimidine tract in large introns, are essential for splicing. In addition, several studies have demonstrated the importance of pre-mRNA and mRNA secondary structures on splicing (4–9).
Evidence for the maintenance of pre-mRNA secondary structures has been found in the introns of the alcohol dehydrogenase gene (Adh) in diverse Drosophila species. Schaeffer and Miller (10) used the Adh region in Drosophila pseudoobscura to study epistatic fitness interactions and found two significant clusters of linkage disequilibria in the Adh adult intron and intron 2, although not in intron 1. However, a recent study of the Adh paralogs in cactophilic Drosophila (11) identified two distinct haplotypes in intron 1 of both the Drosophila mojavensis and Drosophila arizonae Adh-2 genes. The observed linkage disequilibria and haplotype structures in these introns are likely to be caused by epistatic selection maintaining pre-mRNA secondary structures (10–12). In addition to the population studies, the region forming a putative hairpin structure upstream of the branch point of intron 1 is found to be very well conserved in the melanogaster subgroup and in the obscura group (12, 13). Similar structures are also found in the subgenus Drosophila and the Hawaiian picture wing group (13). The structure is supported by two independent covariations detected in the complementary sequences of the putative hairpin structure in the genus Drosophila (12). A likelihood ratio test developed by Muse (14) also strongly supports the phylogenetically predicted hairpin structure (12).
In this study, we investigate the effect of this putative hairpin structure on splicing efficiency and protein production. Kimura's compensatory neutral evolution model predicts the following results: if the hairpin structure is involved in splicing and therefore maintained by selection, mutations disrupting the pairing are expected to lower the fitness, whereas compensatory mutations that restore the hairpin structure should have a fitness equivalent to the WT. Using this putative hairpin structure, we also investigate the effect of stem length and secondary structure stability on splicing efficiency and ADH protein level.
Materials and Methods
Target Sites for Site-Directed Mutations. Site-directed mutagenesis was performed to eliminate the entire stem of the putative hairpin structure instead of introducing a single mismatch in the stem. This was done for two reasons. First, single nucleotide substitutions are often observed within the phylogenetically conserved pairing regions, causing a mismatch in the hairpin structure. Even though most of these polymorphisms are at low frequencies within populations, some of them nevertheless rise to intermediate frequencies, such as one haplotype found in a D. pseudoobscura population survey (15). Two substitutions within the Adh intron 1 region reduce the 9-bp stem to 5 bp, yet it is found in 6.43% of all strains and 20% in the Sonoma, CA, population (10, 15). Second, by applying a two-locus model of reversible mutations with compensatory fitness interactions to divergence data from the bcd 3′ UTR secondary structure of the D. melanogaster/Drosophila simulans and D. melanogaster/D. pseudoobscura species pairs, Innan and Stephan (16) estimated that the average selection intensity (Nes) against single deleterious mutations in RNA secondary structures is unlikely to be much >1. This finding suggests that the deleterious effect of a single mismatch is relatively small, and the results of mutational analysis of single nucleotide substitutions in the hairpin structure may not be observable in the laboratory.
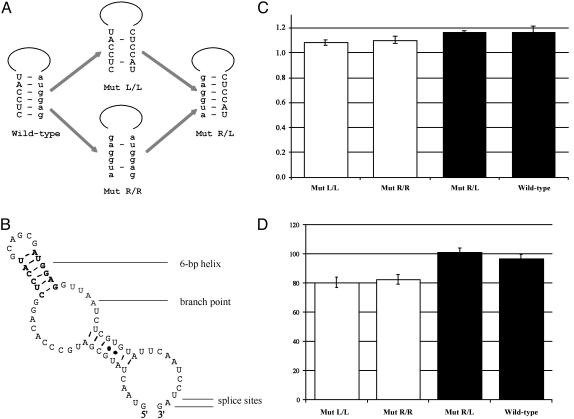
For clarity, the conserved pairing regions 899–904 and 911– 916 of the intron are referred to as L (the left side of the hairpin structure) and R (the right side), respectively. All coordinates are from ref. 17. The Mut L/L mutant construct contains nucleotide substitutions A911C, G913C, G914C, and G916T; Mut R/R contains nucleotide substitutions C899A, C901G, C902G, and T904G; and the double mutant construct Mut R/L contains all eight substitutions in Mut L/L and Mut R/R (Fig. 1A).
Fig. 1.
Mutational analysis of the putative hairpin structure in D. melanogaster Adh intron 1. (A) Mutant constructs used in the experiments. The conserved nucleotide positions 899–904 (L) are shown in capital letters and 911–916 (R) in lowercase letters. A dash between two nucleotides indicates Watson–Crick pairing. (B) RNA secondary structure of D. melanogaster Adh intron 1 predicted by mfold 3.1 (18). The conserved pairing region is in bold, and the branch point sequence (UUAA) is shown in shadow. (C) Means and standard errors of the RSE of the WT and each mutant construct. The constructs for which the conserved secondary structure is disrupted are shown as white columns, whereas the constructs with the secondary structure restored or intact are shown in black. Confidence intervals (95%) are indicated by error bars on each column. (D) Means and standard errors of the ADH enzyme activities of the WT and the mutant transformed lines. The color scheme follows C. Confidence intervals (95%) are indicated by error bars on each column.
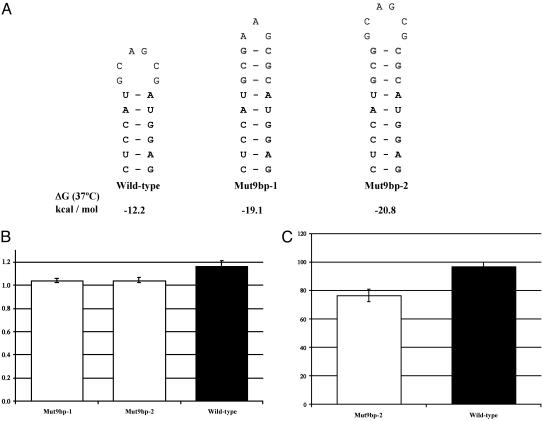
Further experiments strengthening the hairpin structure were carried out to investigate the effect of stability on splicing efficiency. The secondary structure of the WT D. melanogaster Adh intron 1 with the putative 6-bp hairpin structure, as shown in Fig. 1B, has a folding free energy of –12.2 kcal/mol at 37°C, predicted by mfold 3.1 (18). Inserting additional pairings into the hairpin structure stabilizes the entire intron secondary structure. Mutant construct Mut9bp-1 was made by inserting three additional nucleotides (907GA and 911C, coordinates according to the WT sequence) in the loop of the hairpin structure (Fig. 2A). This strengthens the original 6-bp stem to 9 bp and significantly decreases the folding free energy to –19.1 kcal/mol. The branch point and the distance from the putative hairpin structure to the branch point are unaffected by these insertions. To eliminate any effect of the loop size difference between the WT and Mut9bp-1, another mutant construct, Mut9bp-2, was made (Fig. 2 A). Mut9bp-2 has the same nucleotides in the loop as the WT and the same 9-bp stem and similar folding free energy (–20.8 kcal/mol) as Mut9bp-1. Mut9bp-2 has six nucleotides inserted (907GGC and 911CGC) compared with the WT.
Fig. 2.
Mutational analysis of the secondary structure stability. (A) Mutant constructs used in the experiments. The conserved pairing regions are in bold, and the additional pairings in the mutant constructs are shown in shadow. Below each structure is the folding free energy ΔG given by mfold 3.1 (18). (B) Means and standard errors of the RSE of the WT (black) and the 9-bp stem mutant constructs (white). Confidence intervals (95%) are indicated by error bars on each column. (C) Means and standard errors of the ADH enzyme activities of the WT (black) and Mut9bp-2 (white) transformed lines. Confidence intervals (95%) are indicated by error bars on each column.
Construction of Transfection and Transformation Plasmids. All Adh constructs were derived from an 8.6-kb SacI/ClaI genomic Adh fragment of the D. melanogaster Wa-F allele (17). The pUC Wa-F plasmid (kindly provided by J. Parsch, University of Munich) contains the 8.6-kb fragment inserted into pUC18 and was used for site-directed mutagenesis. Mutant clones were sequenced to confirm the mutations and ensure that no other unintentional mutations occurred.
The pUC Wa-F plasmid was used directly in transfection experiments. The control plasmid used in transfection was derived from Adhfn24. This ADH-null allele contains an 11-bp deletion in the Adh exon 2, is transcribed, but has premature termination (19). The size difference of 11 nt in both the pre-mRNA and processed mRNA differentiates the control plasmid from those of the Wa-F-derived plasmids.
For each transformation construct, the appropriate 8.6-kb Adh fragment with the desired mutations was cloned into the YES transformation vector (20). The YES vector is a P element-based vector containing the yellow gene as a selective marker, and the suppressor of Hairywing binding regions as insulators against positional effects (21). The entire Adh coding region in the YES transformation plasmid was again sequenced to confirm the mutations.
Transfection of D. melanogaster Schneider L2 Cells and Comparative Quantitative RT-PCR. D. melanogaster Schneider L2 cells have been reported to not express the endogenous Adh gene (22). This idea was tested and confirmed before the transfection assay. Endotoxin-free supercoiled plasmids (5 μg of the Adhfn24 control plasmid and 5 μg of the Wa-F-derived plasmid) were used to perform transient transfection in Schneider L2 cells (kindly provided by C. Benyajati, University of Rochester) with the calcium phosphate–DNA transfection method (22).
Total RNA was extracted from the transfected cells and treated with RNase-free DNase to eliminate any contaminating genomic or plasmid DNA. This step is extremely important because a large amount of plasmid DNA (10 μg) has been previously added to the cells. The DNA-free total RNA (1 μg) was then reverse-transcribed by using poly(dT) oligonucleotides, and the product was used as template for the subsequent PCR. The pair of primers used to amplify the region containing Adh intron 1 is from 739–762 and 1,134–1,153. To accurately quantify the PCR product, reactions were carried out within the linear range of amplification (<22 cycles).
Because the ratio of mRNA to precursor mRNA levels is a more sensitive indicator of the rate of splicing than the mRNA levels alone (23), amplifications of both mRNA and pre-mRNA were quantified by measuring the intensity of DNA bands ran on a 3% agarose gel stained with SYBR-Gold (Molecular Probes), using imagequant software (Molecular Dynamics). Splicing efficiencies were measured as the ratio of mRNA to the sum of mRNA and pre-mRNA. Relative splicing efficiencies (RSE) were calculated as the ratio of the splicing efficiencies of the Wa-F-derived plasmid (either mutant or WT) to that of the Adhfn24 control plasmid.
P Element-Mediated Germ-Line Transformation of D. melanogaster and ADH Activity Assay. The YES plasmid was used for germ-line transformation (20, 24–26). Germ-line transformation was achieved by microinjection of D. melanogaster y w; Adhfn6; Δ2–3, Sb/TM6 embryos (27). After microinjection, adult flies were crossed to the y w; Adhfn6 stock, and transformed offspring were identified by dark body color (y+). Multiple independent insertion lines were generated by crossing the transformed flies containing both the YES insertion and the Δ2–3 source of transposase to the y w; Adhfn6 stock. Transformants with insertions at new locations were identified as dark-bodied (y+) offspring of which the y+ marker was not segregating with the chromosome containing the initial insertion. Only autosomal insertion lines were used for analysis due to dosage compensation of the X chromosome in Drosophila (20, 28). Transformed lines containing multiple Adh insertions were identified by Southern blotting and excluded from further analysis.
ADH enzymatic activity was measured by the spectrophotometric method (29) using isopropanol as a substrate. Four males from each transformed line were crossed to four females of the y w; Adhfn6 stock, and 10 male progeny heterogeneous for the Adh insertion were collected at age 6–8 days and used for the ADH activity assay. Two separate crosses were made for each transformed line, and two separate assays using five heterogeneous male progeny were performed for each cross, resulting in a total of four measurements for each transformed line. Unit of ADH activity is defined as μmol of NAD+ reduction per min per mg of total protein multiplied by 100. Total protein was estimated by the Folin phenol procedure (30). Differences in ADH activity between the mutant and the WT constructs were tested by a nested ANOVA model that contains the effects of construct type, line within construct type (insert positional effects), cross, replicate within cross, and all of their interactions.
Results
Disruption of the Putative Hairpin Structure in Adh Intron 1 Reduces Splicing Efficiency, Whereas the Compensatory Double Mutant Restores It. To test whether the phylogenetically conserved hairpin structure in the intron is maintained by selection, mutations were introduced into the pairing regions (Fig. 1 A). Mut L/L disrupts the stem of the putative hairpin structure by replacing the sequence on the right side of the stem with that from the left side. Similarly, Mut R/R disrupts the hairpin structure by replacing the left side of the stem with the sequence from the right side. The double mutant construct, Mut R/L, switches the left and right sides of the stem, so that pairing is restored, even though the primary sequence is different. The length of the intron and the rest of the Adh sequence for all three mutant constructs are the same as WT. If the hairpin structure has a functional role in splicing, then the splicing efficiency in both Mut L/L and Mut R/R is expected to deviate from the WT. If only the secondary structure rather than the primary sequence is important, then the double mutant Mut R/L is expected to restore the splicing efficiency to the WT level.
The splicing efficiencies of the three mutant and the WT constructs were assayed in Schneider L2 cells. Each mutant construct was transiently transfected into the culture, together with the control plasmid containing Adhfn24. RSE were measured as the splicing efficiency of each construct standardized to the splicing efficiency of the Adhfn24 control plasmid. The RSE of six transfection assays of the Mut L/L construct has a mean of 1.084 and a standard deviation of 0.029, the RSE of five transfection assays of Mut R/R is 1.103 ± 0.035, and the RSE of six transfection assays of Mut R/L is 1.163 ± 0.018. For comparison, the RSE of six transfection assays of the WT construct is 1.166 ± 0.057 (Fig. 1C). Fisher's significance test was used to examine whether the RSEs of Mut L/L and Mut R/R are significantly different from that of the compensatory mutant Mut R/L and the WT. The tests show no significant differences between the RSEs of Mut L/L and Mut R/R (P = 0.419), or Mut R/L and the WT (P = 0.897). However, all four pairwise comparisons between constructs disrupting the hairpin structure (Mut L/L and Mut R/R) and constructs maintaining it (Mut R/L and the WT) are significant (P values range from 0.001 to 0.016).
These results show that disruption of the putative hairpin structure in D. melanogaster Adh intron 1 decreases the relative splicing efficiency of the intron in Schneider L2 cells, suggesting that the phylogenetically conserved hairpin structure may be functionally important in the splicing process and is maintained by natural selection.
Reduced ADH Activity Is Associated with Disrupted Pairing in the Intron, Whereas the Compensatory Double Mutant Has Similar ADH Activity as the WT. To confirm the effect of the phylogenetically conserved hairpin structure in Adh intron 1 and investigate whether it plays a functional role in a complex multicellular organism, P element-derived YES vectors containing the same mutant constructs as above were introduced into a D. melanogaster Adh– line. The ADH activities of the transformants were measured and compared with the control lines of Adh WT transformants. All transformant lines used in the ADH activity assay are single insertion lines verified by Southern blots on genomic DNA. The mean ADH activity of nine independent Mut L/L transformed lines and nine Mut R/R lines are 80.6 and 82.6 units, respectively (Fig. 1D). Both are significantly lower (P < 0.01) than the mean ADH activity of 12 WT transformed lines (96.7 units), as tested by ANOVA. In contrast, the average ADH activity of 11 independent Mut R/L lines is 101.1 units, significantly higher than that of Mut L/L and Mut R/R lines (P < 0.01), but not significantly different from that of the WT (P = 0.258, Bonferroni's post hoc test). Because all mutations are made in intron 1, and the Adh mRNA and ADH protein of all transformants are identical, the difference in ADH activity must be caused by the difference in ADH protein production. This reflects the difference in the amount of mature Adh mRNA produced, such as found in transfection assays in Schneider L2 cells.
Higher Stability of the Adh Intron 1 Secondary Structure Reduces Splicing Efficiency and Decreases ADH Activity. Mutant constructs that strengthen the phylogenetically conserved hairpin structure in Adh intron 1 (Fig. 2 A) were made to investigate the effect of secondary structure stability on splicing efficiency and ADH activity. The hairpin structure of the WT has a 6-bp stem. This is extended to 9 bp in both the Mut9bp-1 and Mut9bp-2 constructs. Furthermore, Mut9bp-2 has the same 6-nt hairpin loop as the WT. Even though the lengths of the intron in the mutant constructs are slightly increased, the rest of the Adh sequence and the distance between the hairpin structure and the branch point remains the same. The functionally important hairpin structure is also maintained, although with a longer stem length.
The splicing efficiencies of the WT and two mutant constructs were assayed in Schneider L2 cells. The mean RSEs of nine transfection assays of Mut9bp-1 and Mut9bp-2 are 1.044 ± 0.028 (SD) and 1.046 ± 0.034, respectively. For comparison, the mean RSE of six transfection assays of the WT is 1.166 ± 0.057 (Fig. 2B). A two-tailed independent t test gives probabilities of 0.0001 and 0.0002 when the mean RSE of the WT is compared with that of Mut9bp-1 and Mut9bp-2, respectively. The differences are highly significant, indicating that a more stable secondary structure in intron 1 reduces splicing efficiency.
To confirm the results from the transfection assays and investigate the effect of a more stable intron secondary structure on the amount of ADH protein produced, germ-line transformation of Mut9bp-2 was performed. The seven independently transformed Mut9bp-2 lines have a mean ADH activity of 76.5 units, compared with the 12 WT transformed lines of 96.7 units (Fig. 2C). The difference is highly significant (P < 0.001) by ANOVA.
Thus, even though a particular hairpin structure in D. melanogaster Adh intron 1 is preferred and evolutionarily maintained for its role in splicing, a very stable secondary structure in this intron nevertheless causes a reduction in both splicing efficiency and ADH production.
Discussion
Weak Epistatic Selection Maintains the Phylogenetically Conserved Hairpin Structure in Adh Intron 1. We found that disruption of the hairpin structure causes a small reduction (6.1%) in splicing efficiency of intron 1 (Fig. 1C) and a ≈15% reduction in ADH activity caused by the change in ADH protein level (Fig. 1D). The results are consistent with Kimura's compensatory evolution model: mutations that disrupt the conserved hairpin structure (Mut L/L and Mut R/R) are expected to be deleterious and deviate from the WT in splicing efficiency and ADH protein level; on the other hand, the compensatory mutant, Mut R/L, which restores the hairpin structure and is expected to be neutral, has a splicing efficiency and ADH level not significantly different from the WT.
The hairpin structure we investigated seems to be under weak selection for the following reasons. First, extensive quantitative variation in ADH protein abundance and activity has been observed in natural populations of D. melanogaster, much larger than the range of differences found in our assays (31, 32). However, even for the fast and slow Adh alleles, which when being homozygous lead to 2- to 3-fold differences in ADH activity, the differences in fitness experiments are not significant under noncompetitive conditions (e.g., ref. 33). This finding suggests that the small differences in ADH abundance observed in our experiments are associated with rather small fitness effects.
Second, theoretical models of compensatory evolution have predicted that when selection against deleterious intermediate states is strong (Nes ≫ 1), the rate of compensatory evolution (i.e., the inverse of the transition time from one Watson–Crick base pair to another) is very low (34). However, the observation that evolution in stems of RNA secondary structures is generally only five times slower than in neighboring unpaired regions (16) suggests that selection against deleterious intermediates is weak (Nes ≈ 1). This finding is consistent with the fact that mismatches in stems are found in interspecific sequence comparisons of the Adh intron 1 within the melanogaster subgroup, and covariations occur within the subgenus Sophophora (12).
However, there is one caveat about these arguments in favor of weak selection. Even for the case of strong selection against the deleterious intermediate states, compensatory evolution may still be possible if it happens by template-mediated mismatch repair (35–37). This is a process that increases the mutation rate by allowing concerted mutations on opposite strands of the stem. Because the rate of compensatory evolution is strongly influenced by the mutation rate (16, 34, 38), this would allow the process to occur within a reasonable time scale. However, to test for the possible involvement of template-mediated mismatch repair, one should examine base pairs in an RNA secondary structure that have critical functional roles, such that in introns one mismatch causes a complete splicing defect rather than a small reduction in splicing efficiency. If covariations are still found for those base pairs, then a template-mediated mismatch repair mechanism would seem highly likely.
Does the Conserved Hairpin Structure in Adh Intron 1 Play a Role in Splicing? Because the hairpin structure is physically very close to the putative branch point sequence (UUAA, with the branch point adenosine underlined), it may contribute to splicing efficiency by forcing the downstream branch point sequence into an unpaired conformation (Fig. 1B). It has been shown by compensatory mutation analysis that a highly conserved region in the U2 snRNA forms Watson–Crick base pairs with the branch point sequence, and this bimolecular helix between the U2 snRNA and the branch point sequence is required for the recognition of the branch point and the initiation of splicing (39–42). Thus, the conserved hairpin structure upstream of the branch point sequence may facilitate the base-pairing between the U2 snRNA and the unpaired branch point sequence and increase splicing efficiency.
To test our hypothesis that branch point sequences are unpaired, we tried to identify similar hairpin structures in other short (<100 nt) Drosophila introns by using the phylogenetic comparative RNA secondary structure prediction program, piranah (43). However, because of the lack of a sufficiently large number of interspecific intron sequences that span a moderate phylogenetic range (to allow for enough sequence divergence and possible covariations), only a handful of intron sequence alignments could be used for secondary structure prediction and analysis. We found a possible stem structure in intron 3 of the myosin alkali light chain gene (Mlc1). The putative pairing region is 7 bp long and supported by one covariation in the melanogaster species subgroup. The same pairing region was also predicted by the free energy folding program mfold 3.1 (18) when applied to individual sequences. The putative branch point sequence is located between the two reverse complementary regions such that a pairing of the two 7-nt-long sequences can force the branch point into the unpaired loop region of the hairpin (data not shown). However, a broader phylogenetic range of sequences needs to be analyzed to show that this putative secondary structure is as well conserved as the one found in Adh intron 1 and might play a similar functional role in splicing. In addition, a much larger set of introns is required to further test the hypothesis that branch points are unpaired within pre-mRNA secondary structures.
A Highly Stable Intron Secondary Structure Reduces Splicing Efficiency and Protein Level. Increase in the stability of the intron secondary structure without altering the conserved hairpin structure causes the splicing process to be less efficient than in the WT (≈90% of the WT, Fig. 2B). The result is also confirmed in transformed fly lines, with a reduction in ADH activity of ≈21% (Fig. 2C). In most species groups, a length of ≈6 bp seems to be optimal; in the obscura group, however, the stems are 2–3 bp longer; and in the Hawaiian species, the stem is even longer (13).
More than half of Drosophila introns are <80 nt, and most of them have lengths in the range of 59–67 nt (44). The size of short introns may be important for their recognition; expansion of short introns could inhibit or decrease their splicing both in vitro and in vivo (45–47). The WT D. melanogaster Adh intron 1 is 65 nt in length, and in the two mutant constructs Mut9bp-1 and Mut9bp-2, the length of the intron is increased to 68 and 71 nt, respectively. However, these lengths are still within the range of most Drosophila short introns, so the reductions in splicing efficiency in both Mut9bp-1 and Mut9bp-2 are unlikely to be caused by the slight increase in intron size.
Another possibility is that the reduction in splicing efficiency is caused by a more stable secondary structure of the entire intron. Introns are found to be more A+T rich than their neighboring exons in many diverse eukaryotic organisms (44, 48, 49). The high A+T content of introns may limit their secondary structure stability. Goodall and Filipowicz (50) showed that A+T-rich sequences, in addition to the consensus splice sites, are necessary for the splicing of plant introns. They suggested that A+T-rich sequences or T-rich motifs act as binding sites for heterogeneous nuclear ribonucleoproteins or other splicing factors, and base composition is under selective constraint in plant introns (50, 51). It is also possible that minimal secondary structure formation in introns is beneficial; introduction of hairpin structures between the branch point and 3′ splice site in yeast and mammalian introns may prohibit splicing (52, 53). Computer analyses have also suggested that intron 4 of the Mlc1 gene is under selection for a more open secondary structure (54, 55). The different capacity in forming secondary structures may also help trans-acting splicing factors to differentiate between exons and introns, especially in the recognition of small introns (49, 56). Therefore, the base composition differences in exons and introns could reflect the constraints imposed by secondary structure formation of pre-mRNA. Our result of a reduction in splicing efficiency and protein concentration in the highly stable Mut9bp-1 and Mut9bp-2 constructs is consistent with this hypothesis. However, more introns need to be investigated to see whether the overall stability of intron secondary structures has an effect on splicing efficiency and whether less stable secondary structures of introns are generally preferred.
Acknowledgments
We thank C. Benyajati for providing the Schneider cell line, the Adhfn24 mutant plasmid, and advice on the transfection experiments; J. Parsch for providing the pUC Wa-F plasmid, YES vector, and generous assistance in the lab; J. Baines, D. Carlini, T. Eickbush, J. Fry, T. Platt, and D. Turner for many discussions; and two anonymous reviewers for helpful suggestions on the interpretation of our results. This study was supported by National Institutes of Health Grant GM-58404 and funds from the University of Munich (to W.S.). Y.C. held an Ernst Caspari Fellowship from the University of Rochester.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ADH, alcohol dehydrogenase; RSE, relative splicing efficiencies.
References
- 1.Kimura, M. (1968) Nature 217, 624–626. [DOI] [PubMed] [Google Scholar]
- 2.Kimura, M. (1983) The Neutral Theory of Molecular Evolution (Cambridge Univ. Press, Cambridge, U.K.).
- 3.Green, M. R. (1991) Annu. Rev. Cell Biol. 7, 559–599. [DOI] [PubMed] [Google Scholar]
- 4.Clouet d'Orval, B., d'Aubenton Carafa, Y., Sirand-Pugnet, P., Gallego, M., Brody, E. & Marie, J. (1991) Science 252, 1823–1828. [DOI] [PubMed] [Google Scholar]
- 5.Libri, D., Piseri, A. & Fiszman, M. Y. (1991) Science 252, 1842–1845. [DOI] [PubMed] [Google Scholar]
- 6.Eng, F. J. & Warner, J. R. (1991) Cell 65, 797–804. [DOI] [PubMed] [Google Scholar]
- 7.Goguel, V. & Rosbash, M. (1993) Cell 72, 893–901. [DOI] [PubMed] [Google Scholar]
- 8.Goguel, V., Wang, Y. & Rosbash, M. (1993) Mol. Cell. Biol. 13, 6841–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe, K. J. & Ares, M., Jr. (1997) Proc. Natl. Acad. Sci. USA 94, 12467–12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaeffer, S. W. & Miller, E. L. (1993) Genetics 135, 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matzkin, L. M. & Eanes, W. F. (2003) Genetics 163, 181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirby, D. A., Muse, S. V. & Stephan, W. (1995) Proc. Natl. Acad. Sci. USA 92, 9047–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephan, W. & Kirby, D. A. (1993) Genetics 135, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muse, S. V. (1995) Genetics 139, 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaeffer, S. W., Walthour, C. S., Toleno, D. M., Olek, A. T. & Miller, E. L. (2001) Genetics 159, 673–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Innan, H. & Stephan, W. (2001) Genetics 159, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreitman, M. (1983) Nature 304, 412–417. [DOI] [PubMed] [Google Scholar]
- 18.Zuker, M., Mathews, D. H. & Turner, D. H. (1999) in RNA Biochemistry and Biotechnology, eds. Barciszewski, J. & Clark, B. F. C. (Kluwer, Boston), pp. 11–43.
- 19.Benyajati, C., Place, A. R. & Sofer, W. (1983) Mutat. Res. 111, 1–7. [DOI] [PubMed] [Google Scholar]
- 20.Parsch, J., Tanda, S. & Stephan, W. (1997) Proc. Natl. Acad. Sci. USA 94, 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patton, J. S., Gomes, X. V. & Geyer, P. K. (1992) Nucleic Acids Res. 20, 5859–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benyajati, C. & Dray, J. F. (1984) Proc. Natl. Acad. Sci. USA 81, 1701–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pikielny, C. W. & Rosbash, M. (1985) Cell 41, 119–126. [DOI] [PubMed] [Google Scholar]
- 24.Rubin, G. M. & Spradling, A. C. (1982) Science 218, 348–353. [DOI] [PubMed] [Google Scholar]
- 25.Spradling, A. C. & Rubin, G. M. (1982) Science 218, 341–347. [DOI] [PubMed] [Google Scholar]
- 26.Parsch, J., Stephan, W. & Tanda, S. (1999) Genetics 151, 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberg, D. A., Posakony, J. W. & Maniatis, T. (1983) Cell 34, 59–73. [DOI] [PubMed] [Google Scholar]
- 28.Laurie-Ahlberg, C. C. & Stam, L. F. (1987) Genetics 115, 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maroni, G. (1978) Biochem. Genet. 16, 509–523. [DOI] [PubMed] [Google Scholar]
- 30.Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. (1951) J. Biol. Chem. 193, 265–275. [PubMed] [Google Scholar]
- 31.Maroni, G., Laurie-Ahlberg, C. C., Adams, D. A. & Wilton, A. N. (1982) Genetics 101, 431–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurie-Ahlberg, C. C. (1985) Isozymes Curr. Top. Biol. Med. Res. 12, 33–88. [PubMed] [Google Scholar]
- 33.Izquierdo, J. I. & Rubio, J. (1989) Heredity 63, 343–352. [DOI] [PubMed] [Google Scholar]
- 34.Stephan, W. (1996) Genetics 144, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ripley, L. S. (1982) Proc. Natl. Acad. Sci. USA 79, 4128–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Boer, J. G. & Ripley, L. S. (1984) Proc. Natl. Acad. Sci. USA 81, 5528–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golding, G. B. (1987) in Genetic Constraints on Adaptive Evolution, ed. Loeschcke, V. (Springer, New York), pp. 151–172.
- 38.Kimura, M. (1985) J. Genet. 64, 7–19. [Google Scholar]
- 39.Parker, R., Siliciano, P. G. & Guthrie, C. (1987) Cell 49, 229–239. [DOI] [PubMed] [Google Scholar]
- 40.Wu, J. & Manley, J. L. (1989) Genes Dev. 3, 1553–1561. [DOI] [PubMed] [Google Scholar]
- 41.Zhuang, Y. & Weiner, A. M. (1989) Genes Dev. 3, 1545–1552. [DOI] [PubMed] [Google Scholar]
- 42.Query, C. C., Moore, M. J. & Sharp, P. A. (1994) Genes Dev. 8, 587–597. [DOI] [PubMed] [Google Scholar]
- 43.Parsch, J., Braverman, J. M. & Stephan, W. (2000) Genetics 154, 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mount, S. M., Burks, C., Hertz, G., Stormo, G. D., White, O. & Fields, C. (1992) Nucleic Acids Res. 20, 4255–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo, M., Lo, P. C. & Mount, S. M. (1993) Mol. Cell. Biol. 13, 1104–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talerico, M. & Berget, S. M. (1994) Mol. Cell. Biol. 14, 3434–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo, M. & Mount, S. M. (1995) J. Mol. Biol. 253, 426–437. [DOI] [PubMed] [Google Scholar]
- 48.Salinas, J., Matassi, G., Montero, L. M. & Bernardi, G. (1988) Nucleic Acids Res. 16, 4269–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Csank, C., Taylor, F. M. & Martindale, D. W. (1990) Nucleic Acids Res. 18, 5133–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodall, G. J. & Filipowicz, W. (1989) Cell 58, 473–483. [DOI] [PubMed] [Google Scholar]
- 51.Ko, C. H., Brendel, V., Taylor, R. D. & Walbot, V. (1998) Plant Mol. Biol. 36, 573–583. [DOI] [PubMed] [Google Scholar]
- 52.Halfter, H. & Gallwitz, D. (1988) Nucleic Acids Res. 16, 10413–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, C. W., Patton, J. G. & Nadal-Ginard, B. (1989) Annu. Rev. Genet. 23, 527–577. [DOI] [PubMed] [Google Scholar]
- 54.Leicht, B. G., Lyckegaard, E. M., Benedict, C. M. & Clark, A. G. (1993) Mol. Biol. Evol. 10, 769–790. [DOI] [PubMed] [Google Scholar]
- 55.Leicht, B. G., Muse, S. V., Hanczyc, M. & Clark, A. G. (1995) Genetics 139, 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robberson, B. L., Cote, G. J. & Berget, S. M. (1990) Mol. Cell. Biol. 10, 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]