Abstract
Covalent modifications of histone tails modulate gene expression via chromatin organization. As examples, methylation of lysine 9 residues of histone H3 (H3) (H3-K9) is believed to repress transcription by compacting chromatin, whereas methylation of lysine 4 residues of H3 (H3-K4) is believed to activate transcription by relaxing chromatin. The Drosophila trithorax group protein absent, small, or homeotic discs 1 (ASH1) is involved in maintaining active transcription of many genes. Here we report that in extreme ash1 mutants, no H3-K4 methylation is detectable. Within the limits of our assays, this lack of detectable H3-K4 methylation implies that ASH1 is required for essentially all H3-K4 methylation that occurs in vivo. We report further that the 149-aa SET domain of ASH1 is sufficient for H3-K4 methylation in vitro. These findings support a model in which ASH1 is directly involved in maintaining active transcription by conferring a relaxed chromatin structure.
Chromatin serves as the template for processing genetic information. The dynamic DNA–protein structure of chromatin is influenced by epigenetic modifications on both the DNA and nucleosomal histones. Chromatin can exist in alternate states: a relaxed state (euchromatin) where underlying DNA becomes accessible to transcription factors, and a condensed state (heterochromatin) where underlying DNA is transcriptionally restricted. The functional organization of chromatin structure is in part the result of targeted covalent modifications on the N-terminal tails of the core histones. The histone code hypothesis suggests that combinations of histone modifications format the chromatin for proper gene regulation (1).
Although histone methylation was first described in 1964 (2), its significance for regulating chromatin structure and transcription remained unclear until the discovery of the first histone methyltransferase (HMTase), SUV39H (3), a SET domain-containing protein. Lysine-directed histone methylation is intriguing, because it is involved in mediating both compacted and relaxed chromatin. Histone H3 (H3) can be methylated at lysine residues 4, 9, 27, and 36. Methylation of lysine 9 modulates gene silencing (3), whereas methylation of lysine 4 appears to function as a signal for gene activation (4). It has been proposed that the combination of methylation on the lysine 4 and 9 residues of histone H3 with methylation on the lysine 20 residue of histone H4 may also serve as a signal for gene activation (5).
The SET domain is present in transcriptional regulators that have opposing functions. For example, consider the three Drosophila proteins that originally defined the SET domain: suppressor of variegation 3–9 [SU(VAR)3–9] is involved in heterochromatin-dependent gene silencing (6, 7), enhancer of zeste, is involved in maintaining both activated and repressed transcriptional states (8), and trithorax (TRX) is involved in maintaining gene activation (9). The HMTase function of the SET domain has been implicated in conferring both active and repressed transcriptional states. The SET domain and two adjacent cysteine-rich regions (the preSET and postSET domains) of the transcriptional repressor SU(VAR)3–9, and its homologues are sufficient for lysine 9 residue of H3 (H3-K9) methylation in vitro (3). This H3-K9 methylation indicates that SET domain activity is involved in transcriptional repression. On the other hand, Set9, a human transcriptional activator that has a SET domain but is devoid of the preSET and postSET domains, is capable of methylating H3 at K4 in a SET-dependent manner (10). This H3-K4 methylation shows that SET domain activity can also be involved in transcriptional activation. It appears that methylation of H3 has different effects on transcription depending on which residue is modified; however, the modification on these residues depends in part on the same protein motif, the SET domain.
The Drosophila absent, small, or homeotic discs (ash1) gene was identified by lethal mutations that cause a variety of imaginal disc developmental defects (11). Although ash1 is a member of the TRX group of genes that is involved in maintaining active transcription of homeotic genes (12), the ASH1 protein (13) contains a SET domain that is 32% identical to that of SU-(VAR)3–9 and contains both preSET and postSET domains. The isolated ASH1 SET domain with its preSET and postSET domains has HMTase activity in vitro with specificity for lysines 4 and 9 of H3 and lysine 20 of histone H4 (5). Here, we show that ASH1 is necessary in vivo for essentially all of H3-K4 methylation that is detectable, and that the SET domain alone is sufficient to methylate H3-K4 in vitro.
Materials and Methods
Drosophila Stocks. Drosophila stocks were maintained at 20°C or 25°C on standard cornmeal, molasses, yeast, and agar food containing tegosept and proprionic acid as mold inhibitors. The wild-type strain used was Canton SR. All of the ash1 alleles used were described previously (11, 13, 14). Heteroallelic ash1mutant larvae were generated by combining the amorphic allele (ash122) with each of five other alleles.
Immunofluorescence of Polytene Chromosomes. Third instar salivary glands were prepared as described in Nowak and Corces (15). Rabbit antidimethyl-H3 (lys4) (anti-H3-K4dm), antidimethyl-H3 (lys9) (anti-H3-K9dm), antidimethyl-H3 (lys36), antidimethyl-histone H4 (lys20), antiacetyl-H3 (lys9), and antiacetylhistone H4 (lys12) (Upstate Biotechnology, Lake Placid, NY) were used at 1:35. Chromosomes were examined using a Zeiss Axiophot microscope. A Photometrics cooled charge-coupled device camera (Roper Scientific, Duluth, GA), metamorph imaging software, and photoshop (Adobe Systems, Mountain View, CA) were used to obtain and present images.
Immunoblot Analysis. Third instar salivary glands samples were homogenized, separated on a 7% SDS/PAGE gel, and transferred onto poly(vinylidene difluoride) membranes. Membranes were blocked in 5% milk in TBS-T (Tris-buffered saline with Tween 20) for 1 h, incubated with a 1:5,000 dilution of anti-H3-K4dm (Upstate Biotechnology) in TBS-T for 1 h, washed, incubated in secondary antibody conjugated to horseradish peroxidase (HRP) (rabbit) (Amersham Pharmacia), and washed. HRP-conjugated proteins were detected by using enhanced chemiluminescence (Pierce). Equal loading of samples was judged by protein staining and by presence of a background band.
Generation of GST Fusion Proteins and GST Pull-Down Analysis. The SET domain of ASH1 was PCR amplified and subcloned into the SmaI-XhoI sites of pGex4TI (Amersham Pharmacia). The construct encodes an in-frame fusion of the SET domain (residues 1300–1448) with GST. Recombinant proteins were expressed in the Escherichia coli XL1 blue strain and solubilized in 1.4% sarkosyl/1 mM DTT/0.1 mg/ml lysozyme by sonication. Glutathione Sepharose beads (Pharmacia) were added to the cleared supernatant. Matrix-bound fusion proteins were washed three times in GST-binding buffer (20 mM Hepes·KOH/2.5 mM MgCl2/10% glycerol/1 mM PMSF/1 mM DTT/200 mM KCl/0.08% Nonidet P-40). GST-binding buffer and 0.2 mg/ml core histones (Upstate Biotechnology) were added to beads and incubated at 4°C overnight. After washing the bound proteins three times in GST binding buffer, SDS/PAGE loading buffer was added, and samples were boiled for 5 min. GST-SET bound proteins were analyzed by SDS/PAGE and Coomassie staining.
HMTase Assay. GST-SET fusion proteins were expressed in bacteria and purified as described above. Recombinant H3 (0.2 mg/ml; Upstate Biotechnology) and 1 μM S-adenosyl l-methionine (Sigma) were added to the matrix bound fusion protein and incubated at 37°C for 1 h. After washing the bound proteins three times in methylase activity buffer, SDS/PAGE loading buffer was added, and samples were boiled for 5 min. Methylated histones were detected by immunoblotting with anti-H3-K4dm, antidimethyl-H3 (lys9), or antidimethyl-H3 (lys36).
Results
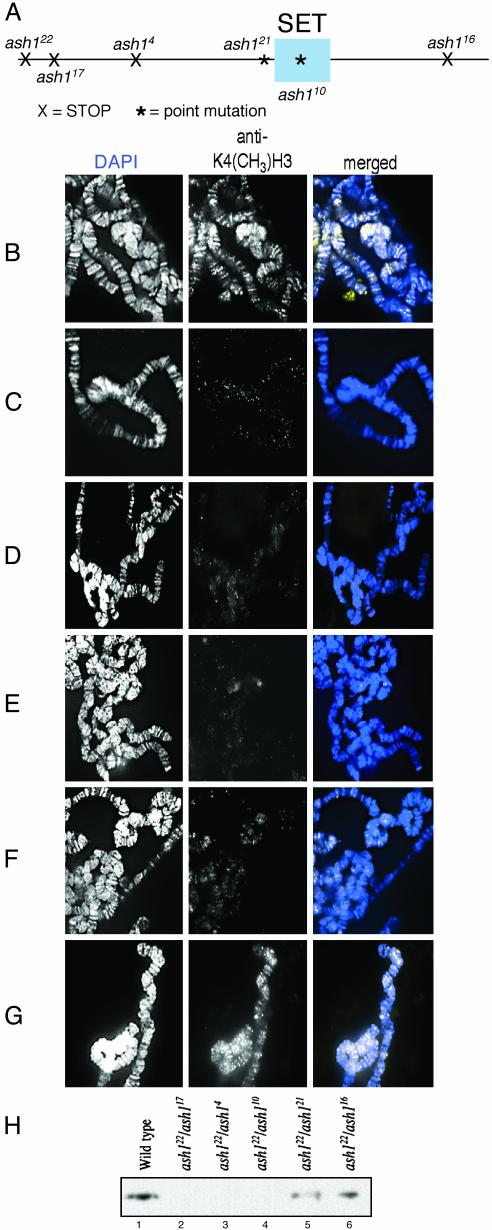
H3-Methylation Is Reduced in ash1 Mutants. Because ASH1 contains a SET domain, and that domain in several proteins had been shown to have HMTase activity, we investigated the HMTase activity of ASH1. In preliminary experiments, we showed that anti-H3-K4dm but not antihistone H1 or H4 can immunoprecipitate ASH1 from Drosophila S2 cells, embryos, and adults (data not shown). This immunoprecipitation indicated that ASH1 is bound to methylated H3-K4 in vivo. Using immunofluorescence, we examined the distribution of histone methylation on polytene chromosomes from ash1 mutant salivary glands. Chromosomes from heteroallelic combinations of the amorphic allele (ash122) with five different mutant alleles were compared with wild-type chromosomes. We previously determined the molecular aberrations of each of the ash1 alleles used (13). A schematic of these aberrations is shown in Fig. 1A. The amorphic allele (ash122) and antimorphic alleles (ash117 and ash14) have early stop codons; their predicted protein products have no preSET, SET, or postSET domains. The missense alleles (ash110 and ash121) have point mutations in the SET domain (N1385I) and in the predicted preSET cysteine-rich region (E1284K), respectively. The weak hypomorphic allele (ash116) has a point mutation leading to a premature stop codon at residue 2001 near the C-terminal end of the protein. In this mutant, the preSET, SET, and postSET domains are intact. Accumulation of ASH1 protein is like wild type on polytene chromosomes from ash110/ash122, ash121/ash122, or ash116/ash122 transheterozygotes(data not shown).
Fig. 1.
Methylation of H3-K4 in ash1 mutants. (A) Schematic showing location of aberrations in ASH1 protein caused by the alleles used in this study. X, STOP codon; *, point mutation. (B–G) Methylation of H3-K4 was detected by immunofluorescence on polytene chromosomes (white, Center; yellowish-white, Right). DNA was detected by 4′,6-diamidino-2-phenylindole (white, Left; blue, Right). (B) Wild type; a large number of euchromatic bands are detected. (C) ash117/ash122, no bands are detected. (D) ash14/ash122, no bands are detected. (E) ash110/ash122, few if any bands are detected. (F) ash121/ash122, some bands are detected. (G) ash116/ash122, like wild-type, a large number of euchromatic bands are detected. (H) Immunoblot of methylated H3-K4 extracted from salivary glands. Lane 1, wild-type salivary glands; lanes 2–6, salivary glands from heteroallelic combinations of ash1 mutations.
Wild-type chromosomes have a widespread distribution of H3-K4 methylation along all of the chromosome arms (Fig. 1B). This H3-K4 methylation is virtually undetectable in either of the antimorphic ash1 mutants (ash117 or ash4) (Fig. 1 C and D). If either truncated protein accumulated at all, it would have no preSET, SET, or postSET domains. The ash110 allele that has a missense mutation in the SET domain also causes undetectable H3-K4 methylation (Fig. 1E). Some H3-K4 methylation is detected at several loci on chromosomes from the ash121 missense allele that has a point mutation in the preSET domain (Fig. 1F). However, there is no detectable reduction in H3-K4 methylation caused by the ash116 missense allele, which has intact preSET, SET, and postSET domains (Fig. 1G). Immunoblot analysis of wild-type and ash1 mutant salivary glands with anti-H3-K4dm confirms these findings showing a significant reduction of H3-K4 methylation in mutants compared with wild type (Fig. 1H). Again, we detected little or no H3-K4 methylation in the antimorphic ash1 alleles, ash117/ash122 (lane 2) and ash4/ash122 (lane 3), and in the missense allele, ash110/ash122 (lane 4). We detected some H3-K4 methylation in ash21/ash122 larvae (lane 5) and close to wild-type levels in ash116/ash122 larvae (lane 6). The remarkable reduction of H3-K4 methylation caused by strong alleles suggests that ASH1 is necessary for essentially all of the H3-K4 methylation that occurs in vivo.
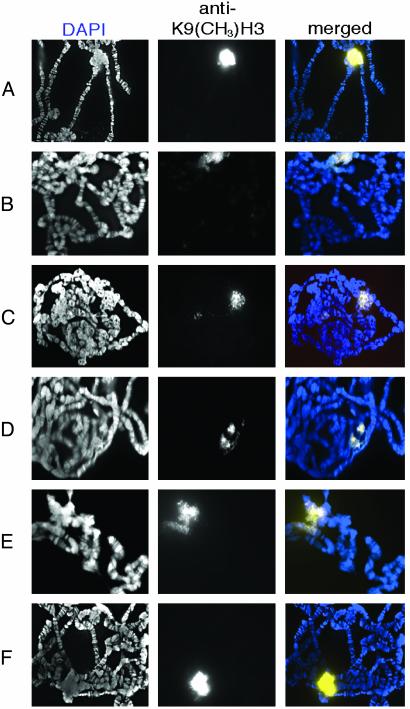
Immunofluorescence with antidimethyl-H3 (lys9) shows that nearly all H3-K9 methylation is detected in the heterochromatic chromocenter of wild-type polytene chromosomes (Fig. 2A) as previously reported (7). In larvae heteroallelic for the antimorphic alleles of ash1 (ash117/ash122 and ash4/ash122), H3-K9 methylation in the chromocenter is slightly reduced compared with wild type (Fig. 2 B and C). In the missense alleles that have a disruption in either the SET domain (ash110) or the preSET domain (ash121), H3-K9 methylation is also reduced (Fig. 2 D and E). By contrast, in an ash1 allele that has intact preSET, SET, and postSET domains (ash116) H3-K9 methylation in the chromocenter is similar to wild type (Fig. 2F). These results suggest that ASH1 may also be required for some H3-K9 methylation.
Fig. 2.
Methylation of H3-K9 in ash1 mutants. H3-K9 methylation was detected by immunofluorescence on polytene chromosomes (white, Center; yellowish-white, Right). DNA was detected by 4′,6-diamidino-2-phenylindole (white, Left; blue, Right). (A) Wild type. (B) ash117/ash122.(C) ash14/ash122.(D) ash110/ash122. (E) ash121/ash122. (F) ash116/ash22.
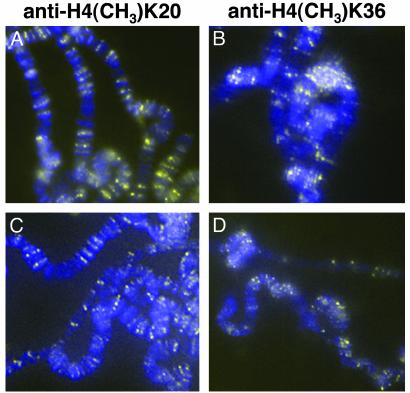
Immunofluorescence with antidimethyl-histone H4 (lys20) (Fig. 3A) and antidimethyl-H3 (lys36) (Fig. 3C) shows a widespread distribution on chromosome arms from wild-type larvae. In larvae heteroallelic for the antimorphic allele of ash1 (ash14/ash122), the distribution of methylated H4-K20 (Fig. 3B) and methylated H3-K36 (Fig. 3D) is not detectably different from in wild-type larvae. These results suggest that other enzymes are responsible for all or nearly all of the methylation of H4-K20 and H3-K36.
Fig. 3.
Methylation of H4-K20 and H3-K36 in ash1 mutants. Histone methylation was detected by immunofluorescence on polytene chromosomes (yellow). DNA was detected by 4′,6-diamidino-2-phenylindole (blue). (A and B) H4-K20 methylation. (A) Wild type. (B) ash14/ash122.(C and D) H3-K36 methylation. (C) Wild type. (D) ash14/ash122.
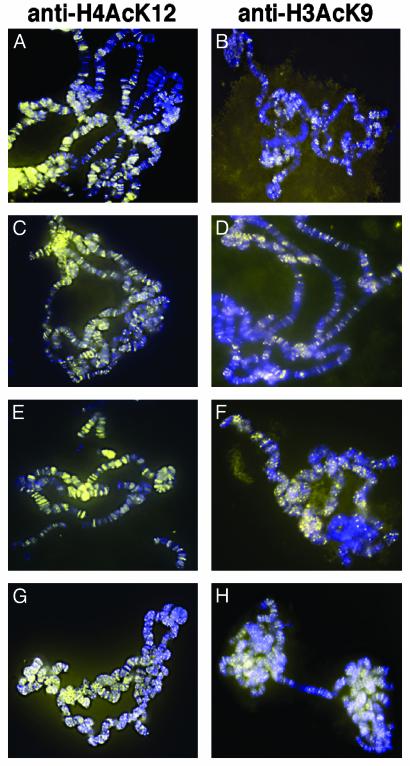
One possible interpretation of the dramatically reduced H3-K4 methylation and slightly reduced H3-K9 methylation we observe is that the ash1 mutants nonspecifically affect the quantity and/or quality of chromosomal histones. That we observed no changes in the pattern of H4-K20 or H3-K36 methylation argues against this interpretation. To address this issue further, we examined acetylation of histones on chromosomes from ash1 mutants. Immunofluorescence with antiacetyl histone H4 (lysine 12) and antiacetyl H3 (lysine 9) from wild-type larvae shows widespread distribution throughout the chromosome arms (Fig. 4 A and B, respectively). This distribution is unchanged in ash1 mutants. There is no reduction in histone H4 acetylated on lysine 12 detected on polytene chromosomes in ash117/ash122 (Fig. 4C), ash14/ash122 (Fig. 4E), and ash110/ash122 (Fig. 4G) mutants, nor is there reduction in H3 acetylated on lysine 9 from in ash117/ash122 in ash117/ash122 (Fig. 4D), ash14/ash122 (Fig. 4F), and ash110/ash122 (Fig. 4H) mutants. Identical results were seen in ash117/ash122 and ash121/ash122 mutants (data not shown). The acetylation levels at lysine 14 of H3 were also unchanged in ash1 mutant nuclei (data not shown). These results suggest that histone integrity is maintained in ash1 mutants and further that loss of ASH1-catalyzed methylation does not affect histone acetylation, at least at the residues examined.
Fig. 4.
Histone acetylation in ash1 mutants. Histone acetylation was detected by immunofluorescence on polytene chromosomes (yellow). DNA was detected by 4′,6-diamidino-2-phenylindole (blue). (A, C, E, and G) Histone H4 acetylation (lysine 12). (A) Wild type. (C) ash117/ash122. (E) ash14/ash122. (G) ash110/ash122. (B, D, F, and H) H3 acetylation (lysine 9). (B) ash117/ash122. (F) ash14/ash122. (H) ash110/ash122.
The ASH1 SET Domain Is Sufficient for Methylation of H3. Recent studies on HMTases suggest that the SET domain in various combinations with its neighboring preSET and postSET domains may play a role in determining catalytic activity and substrate specificity (3, 10, 16). An ASH1 fragment of 588 aa containing the preSET, SET, and postSET domains (residues 1032–1619) is capable of methylating H3-K4 in vitro (5). We examined whether the 149-aa SET domain (residues 1300–1448) by itself can associate with H3 and methylate H3-K4 residues.
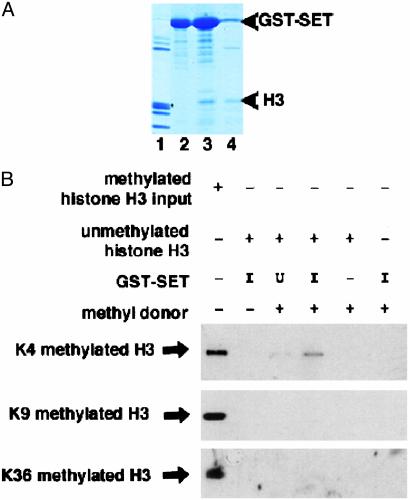
If the role of the ASH1 SET domain is to methylate H3, it should bind directly to H3. Core histones were incubated with bacterially expressed GST-SET fusion protein. The GST-SET fusion protein preferentially pulled down H3 (Fig. 5A, lane 3). H1, H2A, H2B, and H4 were only weakly retained by the recombinant GST-SET fusion protein (Fig. 5A, lane 3). No H3 was detected in the GST-SET input lane (Fig. 5A, lane 2). The leaky expression of uninduced GST-SET was also sufficient to pull down some H3 (Fig. 5A, lane 4). These data show that the SET domain of ASH1 can bind directly to H3.
Fig. 5.
The SET domain of ASH1 can bind to H3 and methylate K4 residues. (A) Partially purified GST-SET fusion protein was used for GST pull-down with core histones. Lanes 1 and 2 show input histones and input GST-SET, respectively. Lane 3 shows GST pull-down with induced GST-SET and histones. Lane 4 shows GST pull-down with uninduced GST-SET and histones. Note that although some nonspecific bands are evident in both the GST-SET input and GST-SET-induced lanes (lanes 2 and 3), none of the bands in the input lane (lane 2) correspond to the molecular weight of H3, which is marked by an asterisk. Upper arrowhead indicates the size of the GST-SET fusion protein; Lower arrowhead indicates the size of H3. (B) To assay for HMTase activity, the partially purified GST-SET fusion protein was incubated with unmethylated H3 and a methyl donor and immunoblotted with antiH3-K4 dm. Lane 1, methylated histone input. Lane 2, GST-SET induced plus unmethylated histones. Lane 3, GST-SET uninduced plus unmethylated histones plus methyl donor. Lane 4, GST-SET induced plus unmethylated histones plus methyl donor. Lane 5, unmethylated histones plus methyl donor. Lane 6, GST-SET induced plus methyl donor. Lanes 1 and 2 show that the anti-K4 methylated antibody detects methylated but not unmethylated H3. Lane 5 shows that the methyl donor will not spontaneously methylate H3. Lanes 3 and 4 show the results of HMTase assay in the presence of uninduced (U) or induced (I) GST-SET, respectively. Coomassie staining revealed equal loading of GST-SET fusion protein in each lane (data not shown).
To investigate whether the ASH1 SET domain is sufficient for histone methylation, we performed in vitro HMTase assays. Bacterially expressed GST-SET was incubated with purified recombinant H3 and a methyl donor. Immunoblot analysis with antibodies that detect methylated lysine residues revealed that H3 was methylated on lysine 4 residues in the presence of a methyl donor and GST-SET but was not methylated on lysine 9 or lysine 36 residues (Fig. 5B, lane 4). Methylated H3 was not detected in the input or uninduced lanes (Fig. 5B, lanes 2, 3, and 5). These data demonstrate that the 149-aa SET domain is sufficient in vitro for H3-K4 methylation.
Discussion
The extent of H3-K4 methylation observed by immunofluorescence on polytene chromosomes from ash1 mutants correlates extremely well with that observed by immunoblotting of salivary gland extracts (Fig. 1). Immunofluorescence on polytene chromosomes provides qualitative information about the genomic distribution of H3-K4 methylation in addition to quantitative information about the extent of H3-K4 methylation. By using immunofluorescence on polytene chromosomes from salivary glands and immunoblotting of salivary gland extracts, we show that detectable H3-K4 methylation is essentially eliminated by strong ash1 mutations. This lack of detectable H3-K4 methylation in ash 1 mutants indicates that ASH1 is required for H3-K4 methylation, but it does not indicate whether that requirement is direct. One possibility is that histone integrity is destroyed in ash1 mutants, and the failure to detect H3-K4 methylation is only a secondary consequence. This possibility was ruled out by showing in those same mutants histone acetylation and methylation of other residues is not affected. ASH1 is a component of a multimeric protein complex (17); another possibility is that some other component of the complex is responsible for histone methylation. In the antimorphic alleles where no full-length ASH1 protein can accumulate, the complex might not form and thus prevent that proposed other component from functioning. One argument against this possibility is that in the missense mutant ash110, no methylation of H3-K4 is detected despite the accumulation of normal amounts of full-length ASH1 protein on polytene chromosomes. However, an even stronger argument against this possibility is that fragments of ASH1 can methylate H3-K4 in vitro (ref. 5 and this work), so the evidence is that ASH1 is required directly for virtually all of the H3-K4 methylation that occurs in vivo.
We confirmed the observation that in wild-type polytene chromosomes the vast majority of H3 methylated on lysine 9 residues is located in the chromocenter (7). In SU(VAR)3–9 mutants, there is strongly reduced accumulation of methylated H3-K9 in the chromocenter (7). It is evident, however, that SU(VAR)3–9 is not the sole HMTase with specificity for H3-K9 in Drosophila, because there is still some H3-K9 methylation on chromosomes from Su(var)3–9 null mutants (7). Su(var)3–9 null mutants are viable, affecting only position effect variegation, suggesting that the catalytic activity of SU(VAR)3–9 alone is not sufficient for global gene silencing. We observed that there is also significantly reduced accumulation of methylated H3-K9 at the chromocenter of ash1 mutants. This was surprising, because ASH1 is not known to play any role in heterochromatic gene silencing. Perhaps ASH1 plays a role in gene silencing in combination with SU(VAR)3–9 that has yet to be discovered. In any case, this means that ASH1 is one of at least two enzymes that can catalyze methylation of H3-K9. This is consistent with the recent report (5) that a 588-aa fragment of ASH1 can methylate both K4 and K9 residues of H3 in vitro. However, neither Schotta et al. (7) nor we detected very much H3-K9 methylation outside of the chromocenter on wild-type chromosomes. The pattern of H3-K9 methylation we observed is completely different from the pattern of H3-K4 methylation, which is not consistent with the idea that ASH1 catalyzes the methylation of both K4 and K9 on the same H3 molecules (5). One possible explanation of this discrepancy is that ASH1 catalyzes only a small fraction of the H3-K9 that occurs in vivo, and that the amount of H3-K9 methylation in the chromocenter catalyzed by SU(VAR)3–9, ASH1, and possibly other enzymes overwhelms our ability to detect any on the chromosome arms. We observed a widespread distribution of acetylated H3-K9 on chromosome arms. If ASH1 methylates H3-K9 along chromosome arms, we might have expected that the level of H3-K9 acetylation would increase in ash1 mutants, because the absence of methylation would increase the availability of free H3-K9 residues. However, no such increase was observed. Beisel et al. (5) reported that their 588-aa ASH1 fragment can also catalyze a low level of methylation of H4-K20. We observed a widespread distribution of methylated H4-K20 along the chromosome arms. However, we did not detect any loss of H4-K20 methylation even in the strongest ash1 mutants. If ASH1 catalyzes H4-K20 methylation in vivo, it must catalyze only a small fraction of the total H4-K20 methylation that occurs.
We expected, on the basis of previous work (3, 10, 16), that the SET domain of ASH1 would be important for HMTase activity. Finding that the ash110 allele that has a substitution within the SET domain (N1385I) causes absence of H3-K4 methylation strengthened that expectation. The significance of this conserved asparagine for the HMTase activity of the ASH1 SET domain is underscored by the recent evidence (5) that H3-K4 methylase activity of a 588-aa ASH1 fragment is lost when this same substitution is introduced. Our finding that the ash121 allele that has a substitution within the preSET domain (E1248K) causes reduction but not elimination of H3-K4 methylation suggests that the preSET domain may affect the efficiency of methylation but is not essential for activity. This conclusion is supported by our data showing that the SET domain by itself (residues 1300–1448) can methylate H3-K4 in vitro. However, this conclusion is not supported by Beisel et al. (5), who reported that the H3-K4 methylase activity of a 588-aa ASH1 fragment (1032–1619) is lost when this same substitution (E1248K) is introduced. Beisel et al. (5) also reported that the 588-aa fragment of ASH1 can methylate H3-K9 residues. We find that the 149-aa SET domain contained within that 588-aa fragment cannot methylate H3-K9 residues, suggesting that the preSET and postSET domains add functionality required for H3-K9 methylation.
Lysine residues can be mono-, di-, or trimethylated. Recent evidence suggests that, at least in Saccharomyces cerevisiae, active genes are trimethylated (18). The antibody we have used to detect lysine 4 methylation of H3 was made against a peptide with a dimethylated lysine. It has the highest specificity for peptides with dimethylated lysine residues, but it can also detect peptides with mono- and trimethylated lysine residues (Technical Services, Upstate Biotechnology). Although we have shown that the 149-aa SET domain of ASH1 can methylate H3-K4 in vitro, we do not know the methylation state of the product we detected. The in vivo function of ASH1 may be to mono- and/or dimethylate H3-K4 residues; di- and/or trimethylation may be the function of some other HMTase. If this is the case, then the absence of H3-K4 methylation we have found in extreme ash1 mutants means that the methylation function of ASH1 creates a substrate essential for subsequent methylation. Support for this idea comes from data showing that in ash2 null mutants, the extent of H3-K4 methylation is reduced, but the distribution is like wild-type (M. Cheng and A.S., unpublished work). The human homologue of ASH2 is in a multimeric complex with SET1, a protein shown to have H3-K4 methylase activity (19). If Drosophila ASH2 is also in a complex with a SET1 homologue, then such a complex may be responsible for subsequent methylation of H3-K4 residues initially methylated by ASH1.
Genetic evidence has indicated that ASH1 is a member of the TRX group of proteins that is involved in maintaining active transcription of many genes. The activities of ASH1 and TRX are functionally related. Mutations in ash1 and trx exhibit intergenic noncomplementation (12); ASH1 and TRX colocalize at multiple sites on polytene chromosomes (20), and ASH1 can be coimmunoprecipitated from embryonic nuclear extracts by antibodies that recognize TRX (20). Moreover, TRX accumulation on polytene chromosomes is reduced in an ash1 mutant (21). TRX has been shown to bind to H3 (22) and to associate with a histone acetyltransferase, dCBP (23). These results, together with the results showing that ASH1 functions as an HMTase, suggest a model for the sequential order to ASH1 and TRX association and in turn histone methylation and acetylation. According to this model, ASH1 binds to H3 via its SET domain and methylates K4 residues. The SET domain of TRX recognizes these methylated H3-K4 residues, which could explain the loss of TRX on chromosomes from an ash1 mutant (21). TRX recruits dCBP, which can then acetylate nearby lysine residues. If this model were correct, one would expect that loss of ASH1 catalyzed methylation would secondarily cause loss of acetylation. Our data, however, do not fulfill this expectation. We found in ash1 mutants, where the levels of H3-K4 methylation are barely detectable, the levels of acetylation on both H3 and histone H4 were unchanged as compared with wild type (Fig. 4). Thus, site-specific changes in histone methylation due to disruption of ash1 have no apparent effect on histone acetylation. It could be, however, that we examined only acetylation of residues that do not depend on methylation of H3-K4. Moreover, ash114, the mutant that showed reduced chromosomal TRX (21), has a molecular defect at nearly the same location as ash116 (13). It is likely that ash114 also has a normal level of HMTase activity, which suggests that TRX requires the C-terminal domain of ASH1 rather than its HMTase activity to bind to chromosomes. Further work will be required to understand the molecular basis of the functional relationship between ASH1 and TRX and the roles of other components of the 2MDa ASH1 complex.
Acknowledgments
We thank Keith Byrd and Scott Nowak for technical advice and comments about polytene chromosome squashes and Victor Corces for the use of his fluorescent microscope. We thank Kimberly Owen from Upstate Biotechnology for sending technical data on the specificity of anti-H3-K4dm. We also thank Mimi Cheng, Michelle Beaucher, Charles Na, Jennifer Martin, Cynthia Wagner, and Matt Harley for critically reading and commenting on this manuscript. This work was supported by a grant from the National Institute of General Medical Science.
Abbreviations: ASH1, absent, small, or homeotic discs 1; HMTase, histone methyltransferase; SU(VAR)3–9, suppressor of variegation 3–9; TRX, trithorax; H3, histone H3; H3-K4, lysine 4 residue of H3; H3-K9, lysine 9 residue of H3; H4-K20, lysine 20 residue of histone H4; anti-H3-K4dm, antidimethyl-H3 (lys4).
References
- 1.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 1–45. [DOI] [PubMed] [Google Scholar]
- 2.Murray, K. (1964) Biochemistry 3, 10–15. [DOI] [PubMed] [Google Scholar]
- 3.Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B. D., Sun, Z., Schmid, M., Opravil, S., Ponting, C. P., Allis, C. D. & Jenuwein, T. (2000) Nature 406, 593–599. [DOI] [PubMed] [Google Scholar]
- 4.Strahl, B. D., Ohba, R., Cook, R. G. & Allis, C. D. (1999) Proc. Natl. Acad. Sci. USA 96, 14967–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beisel, C., Imhof, A., Greene, J., Kremmer, E. & Sauer, F. (2002) Nature 419, 857–862. [DOI] [PubMed] [Google Scholar]
- 6.Tschiersch, B., Hoffmann, A., Krauss, V., Dorn, R., Korge, G. & Reuter, G. (1994) EMBO J. 13, 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schotta, G., Ebert, A., Krauss, V., Fischer, A., Hoffman, J., Rea, S., Jenuwein, T., Dorn, R. & Reuter, G. (2002) EMBO J. 21, 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaJeunesse, D. & Shearn, A. (1996) Development (Cambridge, U.K.) 122, 2189–2197. [DOI] [PubMed] [Google Scholar]
- 9.Capdevila, M. P. & Garcia-Bellido, A. (1981) Roux's Arch. Dev. Biol. 190, 339–350. [DOI] [PubMed] [Google Scholar]
- 10.Nishioka, K., Chuikov, S., Sarma, K., Erdjument-Bromage, H., Allis, C. D., Tempst, P. & Reinberg, D. (2002) Genes Dev. 16, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shearn, A., Hersperger, E. & Hersperger, G. (1987) Roux's Arch. Dev. Biol. 196, 231–242. [DOI] [PubMed] [Google Scholar]
- 12.Shearn, A. (1989) Genetics 121, 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripoulas, N., LaJeunesse, D., Gildea, J. & Shearn, A. (1996) Genetics 143, 913–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripoulas, N. A., Hersperger, E., LaJeunesse, D. & Shearn, A. (1994) Genetics 137, 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowak, S. J. & Corces, V. G. (2000) Genes Dev. 14, 3003–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tachibana, M., Sugimoto, K., Fukushima, T. & Shinkai, Y. (2001) J. Biol. Chem. 276, 25309–25317. [DOI] [PubMed] [Google Scholar]
- 17.Papoulas, O., Beek, S. J., Moseley, S., McCallum, C. M., Sarte, M., Shearn, A. & Tamkun, T. W. (1998) Development (Cambridge, U.K.) 125, 3955–3966. [DOI] [PubMed] [Google Scholar]
- 18.Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Berstein, B. E., Tolga Emre, N. C., Schreiber, S. L., Mellor, J. & Kouzarides, T. (2002) Nature 419, 407–411. [DOI] [PubMed] [Google Scholar]
- 19.Wysocka, J., Myers, M. P., Laherty, C. D., Eisenman, R. N. & Herr, W. (2003) Genes Dev. 17, 896–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozovskaia, T., Tillib, S., Smith, S., Sedkov, Y., Rozenblatt-Rosen, O., Petruk, S., Yano, T., Nakamura, T., Ben-Simchon, L., Gildea, J., et al. (1999) Mol. Cell. Biol. 19, 6441–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuzin, B., Tillib, S., Sedkov, Y., Mizrokhi, L. & Mazo, A. (1994) Genes Dev. 8, 2478–2490. [DOI] [PubMed] [Google Scholar]
- 22.Katsani, K. R., Arrendondo, J. J., Kal, A. J. & Verrijzer, C. P. (2001) Genes Dev. 15, 2197–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petruk, S., Sedkov, Y., Smith, S., Tillib, S., Kraevski, V., Nakamura, Y., Canaani, E., Croce, C. & Mazo, A. (2001) Science 294, 1331–1334. [DOI] [PubMed] [Google Scholar]