Abstract
We describe an efficient system for site-selected transposon mutagenesis in maize. A total of 43,776 F1 plants were generated by using Robertson's Mutator (Mu) pollen parents and self-pollinated to establish a library of transposon-mutagenized seed. The frequency of new seed mutants was between 10–4 and 10–5 per F1 plant. As a service to the maize community, maize-targeted mutagenesis selects insertions in genes of interest from this library by using the PCR. Pedigree, knockout, sequence, phenotype, and other information is stored in a powerful interactive database (maize-targeted mutagenesis database) that enables analysis of the entire population and the handling of knockout requests. By inhibiting Mu activity in most F1 plants, we sought to reduce somatic insertions that may cause false positives selected from pooled tissue. By monitoring the remaining Mu activity in the F2, however, we demonstrate that seed phenotypes depend on it, and false positives occur in lines that appear to lack it. We conclude that more than half of all mutations arising in this population are suppressed on losing Mu activity. These results have implications for epigenetic models of inbreeding and for functional genomics.
In many organisms, genome sequencing has refined gene mapping to the nucleotide level, but the association of genetic function with nucleotide sequence remains a significant challenge (1). For example, whereas it is possible to determine gene function by allele replacement in the mouse (2, 3) this procedure is impractical in plants because of the relatively low rate of homologous recombination. An alternative strategy in Drosophila (4) and Caenorhabditis elegans (5) uses active transposons to generate populations (“libraries”) of organisms harboring new insertional mutations. Individuals carrying insertions in a particular gene of interest can then be selected by PCR with a gene-specific primer and a transposon-specific primer applied to pooled DNA samples from the population. Various pooling schemes minimize the number of DNA preparations that must be performed to identify an individual when a positive result is obtained (6–8).
In maize, two similar strategies have been used to generate collections of plants that can be screened for new insertions into genes of interest such as hcf106 (9), an1 (10), or ZAG1 (11). Both schemes used the transposon Robertson's Mutator (Mu) because of its several advantages: Mu has many copies in the genome (12, 13) and causes a high mutation rate of one new allele per 1,000–10,000 plants (14, 15). Mu elements share closely related 0.2-kb terminal inverted repeats, allowing single primers to recognize multiple elements. Further, new insertions tend to be in genic regions (16) unlinked to the original copy (17). In these and other plant transposon collections, insertions are detected by screening DNA isolated from pooled somatic tissues of the generation preceding the archived seed stock.
One problem with this strategy is that multiple Mu autonomous elements are required to generate high levels of germinal activity, but they also generate high somatic activity (18). Somatic insertions are not transmitted to the next generation, leading to false positives. To avoid this problem, we have designed a scheme to inhibit somatic transposition genetically in the tissue from which DNA is extracted. Using a genetic inhibitor, termed Mu inhibitor, we have developed a population of 43,776 plants containing stabilized Mu insertions. The collection forms the basis for a reverse genetics facility that is openly available to the scientific community.
Materials and Methods
Transposon Insertion Site Screening. DNA samples were prepared from dried leaves by grinding the tissue to powder in a Cuisinart DLC processor, extracting 5 ml of powder with 20 ml of urea buffer plus 20 ml of phenol/chloroform, and precipitating with isopropanol (19). The pellets were resuspended in 2 ml of Tris-EDTA buffer and 2 μl of a 20-fold dilution of each sample, ≈50 ng DNA, were used in PCR. Conditions for PCR were: 1× ExTaq buffer, 0.25 mM each dNTP, 1× Q buffer (a solution of betaine from Qiagen, Chatsworth, CA), 0.5 μM each primer, and 0.5 units ExTaq polymerase (PanVera, Madison, WI) per 20-μl reaction. Primers were: Mu53 (GCCTCYATTTCGTCGAATCC), Mu53s (GCCTCYATTTCGTCGAATC), MuEnd23 (TCGTCYATAATGGCAATTATCTC), and Mu58 (CCAWSGCCTCYATTTCGT) for Mu inverted repeats and vp1–114 (GACGGCATGAGTGAAGAGAA), vp1–224(CACGAGCAACCGCGAAAACA), and vp1–615 (GGCCCTGGTGGAAAGAGTA) for the vp1 gene. Reactions consisted of 35 cycles of 94° for 30 sec, 62° for 30 sec, and 72° for 4 min or, where Mu58 and Mu53s were used, 94° for 30 sec, 58° for 30 sec, and 72° for 4 min. Other primers were designed according to sequences submitted to the maize-targeted mutagenesis (MTM) web site (http://mtm.cshl.org/cgi-perl/newreq.cgi) by using the primer3 program (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). In a typical set of nested amplifications, an outermost primer was used with Mu58, the products were diluted 50-fold, and 2 μl was used as a template with the inner primer and Mu53s. Heritable insertions were identified by preparing DNA from 10 F2 seedlings and repeating the PCR assay.
MTM Database (mtmDB) Web Site. The mtmDB web site (http://mtm.cshl.org) is based on the Apache web server (www.apache.org) and hosts a set of Perl scripts (20) running under the embedded Perl interpreter mod_perl. The underlying database is AceDB (www.acedb.org), which communicates with the web server via the AcePerl library (21). The web interface also depends on the CGI.pm module (22) and the GD module (http://stein.cshl.org/WWW/software/GD).
Genetic Analysis. Ears were scored before shelling, and kernel phenotypes were scored if they segregated on the ear. Parental mutant phenotypes were exhibited by related F1 plants and were not included in the analysis. Approximately 24% and 17% of 1998a and 1998b F2 families had parental mutant phenotypes, respectively (Table 1). Correlation with Mu activity was assessed by using a χ2 contingency test. As a control, correlation with the bz1 sh1 marker genes present in the Mu active lines was also assessed and was significant at the 1% level for total seed mutants in the 1998 populations, possibly reflecting linkage of parental mutations to the bz1 sh1 chromosomal region. Correlations of new kernel mutants with Mu activity were typically significant at P values from 10–6 to 10–12.
Table 1. Mutant and insertion frequency distributions among Mu activity classes.
| 1998a
|
1998b
|
1999
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mu on | Mu off | Mu unscorable* | Total | Mu on | Mu off | Mu unscorable | Total | Mu on | Mu off | Total | |
| Total ears | 2,027 | 3,747 | 4,652 | 10,426 | 4,300 | 903 | 6,930 | 12,133 | 8 | 18,963 | 18,971 |
| Kernel mutants | 1,160 | 1,705 | 2,126 | 4,991 | 1,297 | 200 | 1,789 | 3,286 | 3 | 683 | 686 |
| Frequency, %† | 57 | 46 | 46 | 48 | 30 | 22 | 26 | 27 | 38 | 3.6 | 3.6 |
| Nonparental kernel mutants‡ | 72/192 | 170/805 | 230/942 | 472/1,939 | 275/2,460 | 39/613 | 458/4,566 | 772/7,639 | 3 | 683 | 686 |
| Frequency, % | 38∥ | 21∥ | 24 | 24 | 11∥ | 6.4∥ | 10 | 10 | 38∥ | 3.6∥ | 3.6 |
| Nonparental vp mutants§ | 24 | 37 | 70 | 131 | 6 | 1 | 18 | 25 | 0 | 17 | 17 |
| Frequency, % | 1.2 | 1.0 | 1.5 | 1.3 | 0.14 | 0.11 | 0.26 | 0.21 | 0 | 2.5 | 0.09 |
| Genic insertions¶ | 12/2,027 | 18/3,747 | 22/5,746 | 52/11,520 | 8/4,300 | 0/903 | 8/4,013 | 16/9,216 | 0/23,040 | 1/23,040 | 1/23,040 |
| Frequency, % | 0.59 | 0.48 | 0.38 | 0.45 | 0.19 | 0 | 0.20 | 0.17 | 0 | 0.004 | 0.004 |
Ears in which Mu activity could not be scored because the F1 individual inherited the bz1 sh1 linkage group rather than the bz1-Mum9 reporter gene.
Frequency of indicated mutant type within each Mu subclass.
Kernel mutants were deemed parental if scored in sibling plants at near-Mendelian ratios. The class “nonparental mutants” was defined stringently to comprise aggregate kernel data less all data derived from Mu parents that showed evidence of harboring parental mutants in any branch of the pedigree. The numerator indicates number of mutants, and the denominator indicates the subpopulation size.
Nonparental mutants that displayed a viviparous kernel phenotype.
Mu insertions into user-submitted genes recovered by PCR-based methods. Because tissue grid and ear populations do not coincide exactly, numerator indicates number of knockouts, denominator the subpopulation sizes.
Adjacent frequencies that differ significantly at the 0.5% level.
Results and Discussion
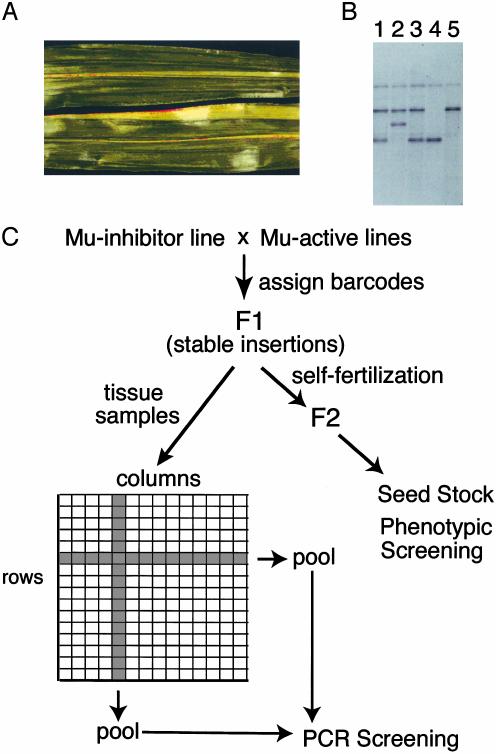
Mutagenesis Scheme and Population Design. It has been previously suggested that somatic insertions of transposon Mu might be responsible for clonal homozygous mutant sectors in leaf tissue, also known as Mu stripes (14). Mu stripes can occupy as much as 1/8th leaf width (Fig. 1A) but are rarely transmitted to the next generation (data not shown). To determine whether these stripes were caused by somatic insertions, DNA was isolated from pale green stripes found in mature hcf106/+ plants and from adjacent tissue. DNA gel blots (Fig. 1B) and amplification and sequencing (data not shown) revealed that more than half of these stripes had new Mu insertions at the Hcf106 locus. Serial dilution of sector DNA with WT DNA revealed that 1 part in 256 could be readily detected by PCR, confirming that these sectors would result in false positives and defining the maximum pool size for selection of germinal insertions (9, 10).
Fig. 1.
Scheme for selecting germinal insertions of Mu insertions. (A) Somatic transposition of Mu elements results in clonal pale green sectors of homozygous mutant tissue in hcf106/+ heterozygous plants. (B) Genomic DNA was prepared from the pale green sector shown in A (lane 2), normal tissue on either side (lanes 1 and 3), homozygous normal (lane 4), and homozygous mutant (lane 5) plants. DNA was digested with HindIII before DNA gel blot analysis using the hcf106 gene as a probe. Note the insertion of Mu DNA into the lower, WT allele. (C) Pooling scheme to identify germinal insertions. To avoid detecting somatic insertions such as those in A, Mu-active lines were first crossed to a Mu-inhibitor strain to inactivate Mu. Germinal insertions were then detected in the F1 plants. An upper leaf from each plant was deribbed, and opposite halves were used for row and column pools. Because clonal sectors have not been observed to cross the midvein (A), somatic insertions should not appear in row and column pools.
To avoid somatic insertions, active Mu lines (23) were crossed to a line carrying Les28, a dominant lesion mimic mutation (Fig. 1C). This line also carries dominant sectoring factors that inhibit Mu activity (24). When these plants were pollinated by using Mu-active parents, progeny initially retained Mu activity, but lost it during later development. The Les28 phenotype depends on Mu activity, so that phenotypic lesions are visible in the lower leaves where Mu is active, but not in the upper leaves where Mu has been silenced, greatly reducing the frequency of somatic insertions. Progeny of such plants usually lost activity altogether and so resembled those of Mu killer strains described by Freeling and coworkers.∥ Because we do not know whether the factors are the same, we refer to ours as Mu inhibitor.
Each F1 plant was given a five-digit barcode that remained associated with its tissue sample and F2 progeny. For DNA preparations, two upper (i.e., Mu-inactive) leaves were harvested and split into halves, discarding the midvein. As clonal somatic sectors are not observed to cross the midvein, the two samples should never include the same sector, providing additional insurance against false positives. Tissue samples were dried and then arranged in 48-by-48 plant grids. Barcodes were scanned to facilitate tracking the positions of individuals within the grid. Leaf halves constituting each row and column were pooled, and DNA was prepared from the pooled tissue. Each grid of 2,304 plants was therefore represented by 96 DNA samples and an individual plant could be recognized by its unique row and column address (9). Additionally, a small leaf sample from each individual was stored in the well of a microtiter dish for later use. F2 seed were harvested and stored. In 1998, two populations of plants were processed into grids: 11,520 were the progeny of Mu-active lines and the Mu-inhibitor line, whereas 9,216 were the progeny of Mu-active lines and the inbred line B73. These plants were arranged into nine grids. In 1999, an additional 23,040 plants were processed from Mu-active and Mu-inhibitor parents and arranged into 10 grids.
Transposon Activity and Mutant Phenotypes. To monitor Mu activity, Mu-active parents of the 1998 planting carried bz1-Mum9 (23), whereas parents of the 1999 planting carried a1-Mum2 (18), both of which spotted seeds only in the presence of Mu activity (Table 1). The Mu-inhibitor strain was the same in each case. In the 1998 planting, 35% [2,027/(2,027 + 3,747)] of F1 plants crossed with Mu inhibitor had F2 progeny that retained Mu activity, but in the 1999 planting there were virtually none (6 of 18,971 scored). In comparison, 83% [4,300/(4,300 + 903)] of F1 plants from crosses of Mu-active lines to B73 still had Mu activity. We conclude that the Mu-inhibitor line dominantly inactivated Mu depending on the Mu line used.
Similar phenotypes in different F2 families from related F1 parents were assumed to be parental mutations in the first planting (1998), and such parental mutations were avoided as much as possible in the second planting (1999). The frequency of nonparental F2 phenotypes is shown in Table 1. For example, nine new sugary mutants were recovered. As there are six known sugary loci, this finding indicates an allele frequency of 1.5 (or 3.4 × 10–5 per plant) and a 78% probability of recovering an insertion into any given gene. In contrast, 15 viviparous loci are known, and 173 mutants were observed, giving an allele frequency of 12.5. If other, uncharacterized genes exist that give sugary or viviparous phenotypes the frequencies may actually be lower. Allelism of the mutants was not tested, so the numbers could reflect multiple insertions in just a few genes. Indeed, given the disparity between the sugary and viviparous frequencies it is likely that Mu has hotspots for insertion into one or more vp genes, and that mutation frequencies may vary between 10–4 and 10–5 per locus.
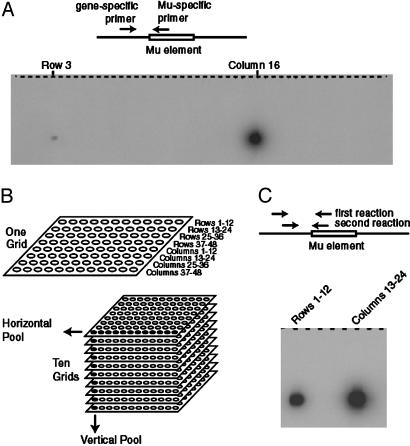
Molecular Selection of Insertions via PCR. Initially, screening was attempted by using PCR between a gene-specific primer and a Mu-specific primer (9). The viviparous1 gene (vp1) was used as a test case because it is the only known viviparous mutation that also eliminates kernel pigmentation (25, 26) and a single viviparous, anthocyaninless mutant was observed in the collection. As shown in Fig. 2A, amplification of pooled DNA from the row and column corresponding to the vp1 mutation was successful, providing a unique address for the new allele. This finding indicated that new insertions could be detected, but the sensitivity was low and, at this level of pooling, 1,824 reactions would be needed to screen the entire collection. We increased the sensitivity by using sequential amplifications with nested primers and reduced the number of reactions by pooling the DNA samples. Each grid of 96 pools (48 row pools and 48 column pools) was pooled horizontally into 8 superpools, and each grid position was pooled vertically across 10 grids (Fig. 2B). The increased sensitivity provided by nested PCRs (Fig. 2C) allowed the detection of the vp1 insertion even when DNA from 576 individuals was present in a single reaction. With the information provided by horizontal and vertical pools, an individual carrying a particular insertion can theoretically be identified from among the 43,776 individuals with only 688 reactions.
Fig. 2.
Screening for Mu insertions in Vp1. (A) Simple PCR between a gene-specific primer and a Mu-specific primer. The products were blotted and probed with a fragment of Vp1 generated by primers vp1–114 and vp1–615. Autoradiography was 16 h. Only 48 of the 96 reactions required to screen a grid are shown. (B) Pooling strategy to reduce the number of reactions required to screen the collection. Groups of 12 rows or columns were pooled horizontally across a grid; grid positions were pooled vertically across grids. (C) Detection of the Vp1 insertion in the horizontally pooled pools. Reaction products were blotted and probed as above, except autoradiography was 2 h.
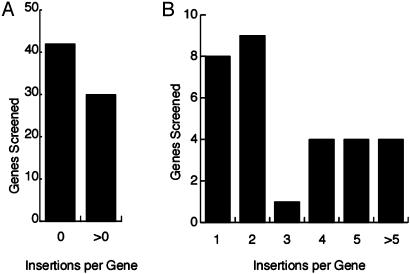
Once the system was demonstrated to work for a test case, a public service was established to allow researchers to obtain mutations in genes of interest. Genomic sequences corresponding to individual genes are submitted via the mtmDB website (http://mtm.cshl.org). The service designs primers to identify insertions throughout the gene and screens the collection. For the first 72 gene sequences submitted 30 had at least one insertion (Fig. 3). Excluding insertions that may have been present in the parents, a total of 65 insertions were found for a mean frequency of 2.1 × 10–5 new insertions per gene per plant. Most insertions (92%) were found in the bz1-Mum9 population from 1998, implying a frequency of 4.0 × 10–5 for these grids. The difference between the 1998 and 1999 populations lies in the different Mu-active parents and underscores that germinal activity of Mu can differ substantially even if somatic activity, scored here by aleurone spotting, appears similar (17). The number of insertions detected molecularly was more consistent with the number of su alleles than vp alleles, again suggesting insertional hotspots in vp. The suggestion of hotspots is supported by the apparent non-Poisson distribution of numbers of insertions per gene (Fig. 3B). Reasons for failure to recover insertions in a given gene include failure of germinal transmission through the pollen, insertional target preference, sampling bias, and incomplete screening caused by submission of partial sequences. If, as seems evident from these data, Mu has target preferences, then complementary mutagenesis programs with other transposons such as Ac will be necessary to recover tagged mutations in all maize genes.
Fig. 3.
Distribution of Mu insertions in 72 genes among 43,776 F1 plants. The insertions were detected by nested PCRs on DNA pools from F1 leaf tissue, and transmission to the F2 was confirmed in all cases by similar PCR analysis on DNA from 1-week-old seedlings.
Unexpectedly, only ≈36% (97/271) of the insertions we detected in F1 tissue were transmitted to the F2 progeny. Of the insertions that were not transmitted, 68% came from F1 plants in which Mu was scored as inactive. For the F1 insertions scored, PCR products were recovered from three independent tissue samples. Thus, it would seem unlikely that many F1 positives were PCR artifacts. Nor does it seem likely that such a majority of mutations created by Mu could not be transmitted. One explanation for the lack of transmission may be that somatic activity of Mu is still occurring but is not reflected in Les28 or kernel spotting. Additionally, the light and temperature dependence of lesions in Les28 plants may complicate phenotypic scoring (24, 27).
The lengths of PCR products observed ranged up to 6 kb. The average size of a coding region in maize is 4.8 kb (ref. 28 and L.S., unpublished work) so that sequences submitted were screened for insertions across a continuous region. For the insertions recovered, the coding regions of each gene were divided in half, and insertions were mapped either by sequence analysis (for genomic sequences) or according to PCR product size (for cDNA sequences). Analysis of 24 insertions indicated a pronounced bias of Mu transposition into the 5′ half of each gene (Table 2), in agreement with anecdotal analysis of a number of maize genes for which multiple Mu alleles have been recovered (9, 29). This preference may account for the high number of suppressible phenotypes observed (see below). Insertions were equally likely to be found in introns or exons, given that maize contains ≈1.5-fold more intron sequence than exon sequence (ref. 28 and L.S., unpublished work). Promoters and 3′ UTRs were not uniformly screened because many submissions did not contain these sequences.
Table 2. Distribution of sequenced insertions in regions of genes.
| 5′ Half | 3′ Half | Intron | Exon | Promoter | 3′ UTR |
|---|---|---|---|---|---|
| 18 | 6 | 11 | 7 | 4 | 1 |
Suppression of Mutant Phenotypes. The phenotypic severity of some transposon-induced alleles can be suppressed in the presence or absence of an active transposon elsewhere in the genome (30, 31). Suppression does not involve excision of the element, but rather the regulatory interaction of transposase with insertions mostly in introns and promoters (32–35), but also in 3′ UTRs (36) (reviewed in ref. 37). We set out to measure the frequency of suppressible phenotypes in MTM, by first eliminating portions of the pedigrees that contained parental mutations. We then investigated the occurrence of suppressible mutations in the F1 by examining the distribution of new F2 kernel mutations with respect to Mu activity. As these mutations must have arisen in the pollen of active Mu parents, Mu activity could only have been lost in the F1 plants.
The frequency of new kernel mutants failed contingency χ2 tests for independence of Mu activity (Table 1; P values 0.00006, 0.0025, and 0.000028), but satisfied control tests for independence of bz1sh1 (Table 1; P values 0.99 and 0.98). That is, fewer new mutant phenotypes were scored in the Mu-off class than in the Mu-on class for all three populations. As new mutations must have arisen in Mu-active pollen, these data suggest that many new mutations were suppressed in F2 ears that had subsequently lost Mu activity. Interestingly, the high frequency of new vp mutants was independent of both Mu activity and bz1 (Table 1), as if the predicted hotspot was not suppressible (see above).
We considered the possibility that the distribution bias of visible mutants in Mu-active F2 ears reflected variation in parental Mu transposition. For example low-activity parents that gave rise to relatively few new mutants could be the source of most Mu-off progeny, whereas high-activity parents that produced many new mutants could generate most Mu-on progeny. In this scenario, Mu transposition events would be expected to show a distribution bias similar to that observed for visible phenotypes. However, the distribution of new Mu insertions detected by PCR screening in F1 plants was independent of Mu activity in the F2, indicating that the frequency of insertions per se was not responsible for the discrepancy in visible phenotypes (Table 1; P values 0.43–0.95).
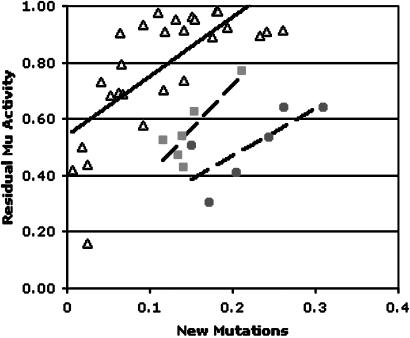
Individual pollen parents contributed both to families with low Mu activity and families with high activity. When the progeny were compared it was possible to correlate activity and mutant frequency directly (Fig. 4). Newly arising phenotypes were positively correlated with Mu activity (Fig. 4, ▵, R = 0.71, P value <0.01). Two sets of six 1998b families each derived from a single Mu parent crossed onto Mu inhibitor showed a similar correlation with residual Mu activity (Fig. 4, ▪, R = 0.85, P value <0.05; Fig. 4, •, R = 0.61, P value <0.10). These data demonstrate that mutants are detected several times more frequently in fully active Mu lines than in fully inactivated lines.
Fig. 4.
Mu dependence of newly arising kernel phenotypes. Each point represents data from a single F1 family, and associated linear regression trend lines are shown. ▴, 1998b population; ▪, six families derived from a single parent, 1998a population; •, six families derived from a single parent, 1998a population.
In summary, germinal transposition of Mu elements in the F0 was independent of presence of Mu activity in the F2, but visible phenotypes were not, implying the existence of a large class of suppressible mutations.
mtmDB Database. To maintain the information regarding the maize Mu population described above and manage the reverse genetics screening process, we designed and implemented a web-based database, mtmDB. mtmDB is based on the AceDB database management system, which is used to curate the C. elegans genomic annotations and a number of plant databases. It describes each of the maize Mu insertion mutants and associated information. It also serves as a public resource for the functional study of maize genomics and as a workflow system for accessing the screening service (http://mtm.cshl.org/flow.html).
Researchers are invited to screen DNA sequences of interest against the collection to identify insertional mutations. If the strain(s) containing an insertion in the DNA segment is already known, associated phenotypic information and seed stocks can be obtained. If a search of the database reveals no match to the sequence of interest, the biologist can send the genomic sequence to Cold Spring Harbor Laboratory for screening by PCR. MTM users are obliged to contribute phenotype information back to the database or indicate that no phenotype was identified. In the long term, such user contributions will help to create a resource for functional genomics in maize.
The web site provides several search options. The main categories are text search, simple search, class search, and advanced search. The text search was designed to emulate the type of search familiar to biologists from web search engines and supports multiple keywords or phrases via the standard and, or, and not boolean operations. The simple search provides name-based access to the biologically most relevant object classes, such as individual plants, grid addresses, and genes. The class search is similar to this, except that it provides access to all of the classes in the database, including ones that are normally not of direct interest, such as Parent_cross. The advanced search is of use to those who know the ACeDB query language and understand the mtmDB data model. This page allows users to pose complex ad hoc queries directly to the database. The data model can be browsed online, and documentation on the ACeDB query language is available (www.acedb.org/Cornell).
The blast search will perform a blastn or tblastn search against mutant sequences contained in the mtmDB database. The intent is to allow users to blastt genes of interest against the database to identify mutants that have already been screened. Because we do not currently make an effort to exhaustively catalog all possible aliases for a gene, it is better to search for a gene by using blast. However, the search page will accept GenBank accession numbers.
Specialized displays exist for each of the major object classes in mtmDB. Some are text based, whereas others use dynamically generated graphics. For queries that retrieve the identity of a mutant strain, the mtmDB web site retrieves and displays an HTML page summarizing all relevant information about the strain, including its phenotype, grid location, and pedigree. Hypertext links inside the data record provide access to more information. These web-page relationships mirror the XREF relationships built into the ACeDB data model. In some cases, it is better to display information graphically. For example, the grid addresses of the mutant and all pedigrees are displayed by default as graphical images. These images are clickable, allowing users to select an element and link to further information retrieved dynamically from the database. Graphical information is also available in a text-only form suitable for cutting and pasting into the researcher's own records.
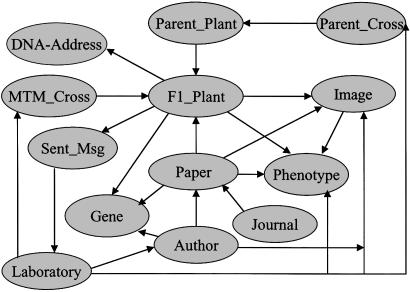
The core of the mtmDB is data concerning Mu insertion mutants, mutant phenotypes, genetic crosses, and specimen tracking information (Fig. 5). The major database classes are as follows. (i) Barcode: plant, seed, and DNA samples from each of the 43,776 F1 lines are labeled with a five-digit barcode. (ii) DNA address: grid positions of barcoded F1 leaf samples, as described later. (iii) Pedigree: crosses used to generate mutants are stored in this class, with different subclasses of pedigree distinguishing the generations. The MTM cross is the final cross used to generate barcoded F1 (currently 279 entries), the parent cross generated the parents and/or grandparents of F1 (currently 44 entries). (iv) Parent plant: the 412 parent and grandparent plants. (v) Phenotype: this class uses a structured vocabulary to describe phenotypic observations. Currently 43,776 ear phenotypes, 8,000 seedling phenotypes, and 2,000 adult phenotypes are in the database. (vi) Image: >700 photographic images. (vii) Laboratory and author. (viii) References: currently 502 papers related to the MTM resource. (ix) Gene: when a line is known to contain a Mu insertion in a described gene it links to this object, which reports the gene name, its possible function, its sequence, and the PCR primers used to detect the insertion. The mtmDB also has a number of classes to manage the registration of researchers with the reverse genetics screening service and track the fulfillment of the user requests.
Fig. 5.
Schema of the mtmDB. Classes of data objects are shown as ellipses, and their interrelationships are represented as arrows.
Conclusions. Mu is the most widely used transposon in maize mutagenesis, both in nontransgenic and transgenic populations (38). The MTM project is a collection of 43,776 maize ears, each containing a unique set of Mu insertions. Phenotypic and genotypic information is accessible at mtmDB (http://mtm.cshl.org), and seeds can be ordered by any academic researcher. The research community is taking advantage of the screening service and will contribute phenotypic information back to the database so that links between maize genomic sequence and gene function can be made.
In addition to its use as a resource, MTM is the largest collection of Mu insertions to be characterized in such detail. The variety of phenotypes is typical of Mu lines, but we have demonstrated a very high proportion of suppressible mutations predicted by our analysis. Such a high frequency of suppressible phenotypes may account for the radical difference in fitness observed between Mu-active and -inactive lines and may have implications for inbreeding depression (39). Suppressible phenotypes can obscure the function of a gene targeted by reverse genetics, but can have utility in mosaic analysis, when examining the cell autonomy of a mutant phenotype (40). By selecting insertions in different locations within the target gene, MTM promises to be a valuable resource in maize developmental biology and maize genetics.
Acknowledgments
We thank Nick Kaplinsky, Phil Becraft, and Matt Evans for screening and identifying kernel phenotypes and Nick Kaplinsky for overseeing kernel phenotype data entry. This work was supported by National Science Foundation Plant Genome Program Grant DBI9872644.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MTM, maize-targeted mutagenesis; Mu, Mutator; mtmDB, MTM database.
Footnotes
Slotkin, R. K., Freeling, M. & Lisch, D. (2003) Maize Genetic Conference Abstracts 45, P209.
References
- 1.Martienssen, R. A. (1998) Proc. Natl. Acad. Sci. USA 95, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee, K. Y., Lund, P., Lowe, K. & Dunsmuir, P. (1990) Plant Cell 2, 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miao, Z. H. & Lam, E. (1995) Plant J 7, 359–365. [DOI] [PubMed] [Google Scholar]
- 4.Kaiser, K. & Goodwin, S. F. (1990) Proc. Natl. Acad. Sci. USA 87, 1686–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zwaal, R. R., Broeks, A., van Meurs, J., Groenen, J. T. & Plasterk, R. H. (1993) Proc. Natl. Acad. Sci. USA 90, 7431–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sussman, M. R., Amasino, R. M., Young, J. C., Krysan, P. J. & Austin-Phillips, S. (2000) Plant Physiol. 124, 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Speulman, E., Metz, P. L., van Arkel, G., te Lintel Hekkert, B., Stiekema, W. J. & Pereira, A. (1999) Plant Cell 11, 1853–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koes, R., Souer, E., van Houwelingen, A., Mur, L., Spelt, C., Quattrocchio, F., Wing, J., Oppedijk, B., Ahmed, S., Maes, T., et al. (1995) Proc. Natl. Acad. Sci. USA 92, 8149–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, L. & Martienssen, R. (1995) Plant Cell 7, 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bensen, R. J., Johal, G. S., Crane, V. C., Tossberg, J. T., Schnable, P. S., Meeley, R. B. & Briggs, S. P. (1995) Plant Cell 7, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mena, M., Ambrose, B. A., Meeley, R. B., Briggs, S. P., Yanofsky, M. F. & Schmidt, R. J. (1996) Science 274, 1537–1540. [DOI] [PubMed] [Google Scholar]
- 12.Chandler, V. L. & Hardeman, K. J. (1992) Adv. Genet. 30, 77–122. [DOI] [PubMed] [Google Scholar]
- 13.Bennetzen, J. L. (1996) Curr. Top. Microbiol. Immunol. 204, 195–229. [DOI] [PubMed] [Google Scholar]
- 14.Alleman, M. & Freeling, M. (1986) Genetics 112, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson, D. S. (1978) Mutat. Res. 51, 21–28. [Google Scholar]
- 16.Hanley, S., Edwards, D., Stevenson, D., Haines, S., Hegarty, M., Schuch, W. & Edwards, K. J. (2000) Plant J. 23, 557–566. [DOI] [PubMed] [Google Scholar]
- 17.Lisch, D., Chomet, P. & Freeling, M. (1995) Genetics 139, 1777–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chomet, P., Lisch, D., Hardeman, K. J., Chandler, V. L. & Freeling, M. (1991) Genetics 129, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, J. & Dellaporta, S. L. (1994) in The Maize Handbook, eds. Freeling, M. & Walbot, V. (Springer, New York), pp. 526–527.
- 20.Wall, L., Orwant, J. & Christiansen, T. (2000) Programming Perl (O'Reilly and Associates, Sebastopol, CA).
- 21.Stein, L. D. & Thierry-Mieg, J. (1998) Genome Res. 8, 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein, L. (1999) The Official Guide to CGI.pm (Wiley, New York).
- 23.Brown, J. & Sundaresan, V. (1992) Genetics 130, 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martienssen, R. & Baron, A. (1994) Genetics 136, 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hattori, T., Vasil, V., Rosenkrans, L., Hannah, L. C., McCarty, D. R. & Vasil, I. K. (1992) Genes Dev. 6, 609–618. [DOI] [PubMed] [Google Scholar]
- 26.McCarty, D. R., Hattori, T., Carson, C. B., Vasil, V., Lazar, M. & Vasil, I. K. (1991) Cell 66, 895–905. [DOI] [PubMed] [Google Scholar]
- 27.Martienssen, R. (1996) in Epigenetic Mechanisms of Gene Regulation, eds. Russo, V. E. A., Martienssen, R. & Riggs, A. D. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 593–608.
- 28.Rabinowicz, P. D., Schutz, K., Dedhia, N., Yordan, C., Parnell, L. D., Stein, L., McCombie, W. R. & Martienssen, R. A. (1999) Nat. Genet. 23, 305–308. [DOI] [PubMed] [Google Scholar]
- 29.Dietrich, C. R., Cui, F., Packila, M. L., Li, J., Ashlock, D. A., Nikolau, B. J. & Schnable, P. S. (2002) Genetics 160, 697–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClintock, B. (1954) Carnegie Inst. Washington Year Book 53, 254–260. [PubMed] [Google Scholar]
- 31.McClintock, B. (1958) Carnegie Inst. Washington Year Book 57, 415–429. [PubMed] [Google Scholar]
- 32.Settles, A. M., Baron, A., Barkan, A. & Martienssen, R. A. (2001) Genetics 157, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barkan, A. & Martienssen, R. A. (1991) Proc. Natl. Acad. Sci. USA 88, 3502–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girard, L. & Freeling, M. (2000) Genetics 154, 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vollbrecht, E., Reiser, L. & Hake, S. (2000) Development (Cambridge, U.K.) 127, 3161–3172. [DOI] [PubMed] [Google Scholar]
- 36.Cui, X., Hsia, A. P., Liu, F., Ashlock, D. A., Wise, R. P. & Schnable, P. S. (2003) Genetics 163, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.May, B. P. & Martienssen, R. (2003) Crit. Rev. Plant Sci. 22, 1–35. [Google Scholar]
- 38.Raizada, M. N., Nan, G. L. & Walbot, V. (2001) Plant Cell 13, 1587–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martienssen, R. (1998) Trends Genet. 14, 263–264. [DOI] [PubMed] [Google Scholar]
- 40.Fowler, J. E., Muehlbauer, G. J. & Freeling, M. (1996) Genetics 143, 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]