Abstract
The p53 gene is a critical tumor suppressor that is inactivated in a majority of cancers. The central role of p53 in response to stresses such as DNA damage, hypoxia, and oncogene activation underlies this high frequency of negative selection during tumorigenic transformation. Mutations in p53 disrupt checkpoint responses to DNA damage and result in the potential for destabilization of the genome. Consistent with this, p53 mutant cells have been shown to accumulate genomic alterations in cell culture, mouse models, and some human tumors. The relationship between p53 mutation and genomic instability in human osteosarcoma is addressed in this report. Similar to some other primary human tumors, the mutation of p53 correlates significantly with the presence of high levels of genomic instability in osteosarcomas. Surprisingly, osteosarcomas harboring an amplification of the HDM2 oncogene, which inhibits the tumor-suppressive properties of p53, do not display high levels of genomic instability. These results demonstrate that the inactivation of p53 in osteosarcomas directly by mutation versus indirectly by HDM2 amplification may have different cellular consequences with respect to the stability of the genome.
The p53 protein is a critical tumor suppressor and central mediator of cellular responses to stress. Inactivating mutations of the p53 gene occur in ≈50% of all sporadic human cancers (1–3). In addition, mutations in known p53-interacting genes such as HDM2 and p14ARF bring the sum total of cancers that display functional inactivation of the p53 pathway to >80% (4–6). Highlighting the central role of p53 in tumor suppression is the predisposition of individuals who inherit a mutated p53 allele to a variety of cancers at an early age including breast carcinoma, soft-tissue sarcomas, brain tumors, osteosarcoma, acute leukemias, and adrenocortical carcinoma (7). Similarly, homozygous p53 knockout mice have a 100% chance of developing cancer (usually thymic lymphomas) before the age of 6 months, and heterozygous mice develop a variety of cancers at a later age including osteosarcoma, soft-tissue sarcomas, and lymphomas (8–11).
The p53 protein mediates responses to a variety of stresses including DNA damage, oncogene activation or mutation, hypoxia, depleted nucleotide pools, shortened telomeres, viral infection, spindle damage, nitric oxide, etc. (12). In response to one or more of these signals, p53 is posttranslationally stabilized and activated as a transcription factor to turn on or off the expressions of different sets of downstream target genes (13). The collective functions of these p53 target genes serve to execute various cellular programs that respond to stress by growth arrest, apoptosis, senescence, or cell–cell signaling (14–16).
Consistent with a role in the response to DNA damage, p53 has been shown to inhibit gene amplifications and deletions, which can be initiated by DNA double-strand breaks. Control over DNA copy number by p53 was first demonstrated by using antimetabolites that induce resistance by gene amplification in cell culture (17, 18). p53 heterozygous (p53+/–) fibroblasts fail to show phosphonoacetyl-l-aspartate-induced amplifications of the CAD gene during culture until loss of heterozygosity (LOH) for p53 (p53–/–), after which gene-amplification frequencies increase by 500- to 1,000-fold. More recently it was demonstrated that these primary human p53+/– cell cultures also undergo spontaneous chromosomal deletions after LOH for p53, as detected by the comparative genome hybridization (CGH) technique, which detects large chromosomal gains and losses in the entire genome (19). A number of reports have shown a similar correlation between the mutational inactivation of p53 and increased genomic instability in vivo. Gene amplifications are common in cells from the normal (nontumor) tissues of the p53 knockout mouse and are detectable as early as 4–6 weeks of age (20). Also, tumors arising in p53 heterozygous mice that undergo LOH for p53 accumulate 5-fold more chromosomal aberrations than tumors in which one wild-type p53 allele is retained (21). In human tumors, mutations in the p53 gene have been shown to correlate with genome-wide instability in colorectal carcinomas (P = 0.05) as defined by CGH (22), with an increase of karyotypic abnormalities and chromosomal amplifications detected by fluorescence in situ hybridization on four chromosomes in breast carcinomas (P < 0.008) (23), and with genome-wide allelic imbalances detected by single-nucleotide polymorphism array analysis in bladder cancer (24). Taken together, these data suggest that the loss of p53 function contributes to tumorigenesis by the destabilization of the genome.
Although the initial experiments describing a role for p53 in surveillance of genomic integrity were performed with primary human fibroblast cell cultures, human sarcomas, which also arise from mesenchymal cells, have not been assayed for genomic instability and p53 status in vivo. Sarcomas, including osteosarcomas and several types of soft-tissue sarcomas, are induced by the loss of p53 function in both heterozygous p53 knockout mice (10, 11) and Li–Fraumeni cancer syndrome patients (7). Also, the effects of mutations in direct p53-interacting genes on genomic stability, such as amplifications of the HDM2 oncogene that occur more frequently in sarcomas than in any other cancer types, have never been examined. The Hdm2 oncoprotein, the gene of which is a downstream transcriptional target of p53, functions as a direct negative regulator of p53 protein function and stability in an autoregulatory feedback loop (25). Whether amplifications of the HDM2 oncogene, which have been demonstrated to impair the tumor-suppressor functions of p53 (26) and occur predominantly in p53 wild-type tumors (27), have an impact on genome stability that mimics that of mutations in p53 has not been established.
One human sarcoma that is ideally suited for analyses of genomic instability is osteosarcoma. Osteosarcomas are the most common malignant bone tumors in adults and children, with a peak incidence during the second decade of life (28). High-grade osteosarcomas are ideally suited for genomic-instability analyses because, on average, these tumors contain as high or even higher levels of genomic instability than almost any other cancer. By CGH, these tumors have ≈9–11 independent chromosomal lesions per tumor, as shown in several previous reports (29–31). In addition to a high mean number of genomic changes, these tumors also collectively display a broad spectrum of instability, where some have ≥20 chromosomal aberrations and others have few or no detectable changes (30), and there is no known genetic rationale for the difference between the two types.
In this report, the role of the p53 tumor-suppressor protein in controlling the stability of the genome in primary high-grade osteosarcomas is addressed. Genomic instability was assayed in 34 primary osteosarcomas by CGH, and the p53 pathway status was determined by both p53 gene mutational screening and genomic copy-number analysis of HDM2. Consistent with the results found with other human cancers, the mutation of p53 significantly correlates with genome-wide DNA instability and seems to represent a major genetic factor contributing to the extremely high levels of genomic instability found in high-grade osteosarcomas. Surprisingly, genomic amplification of the HDM2 oncogene, which occurred independently of mutations in p53, did not correlate with high levels of genomic instability in these tumors. Osteosarcomas with amplified HDM2 displayed less than half as many genomic copy-number alterations on average than those with mutations in p53. These data demonstrate a lack of genetic equivalency for HDM2 oncogene amplifications and p53 tumor-suppressor gene mutations with respect to the stability of the genome in uncultured human tumors.
Materials and Methods
Patients. Osteosarcoma tumor samples were collected from patients who had surgery at Memorial Sloan–Kettering Cancer Center between 1992 and 2000. All samples had a histologically confirmed pathologic diagnosis of high-grade osteosarcoma. All patients provided written informed consent for tissue procurement. Tumors were procured in accordance with a biology study approved by the Memorial Sloan–Kettering Institutional Review Board. Patients from whom these samples came were treated on or according to the pediatric intergroup phase III clinical trial (CCG no. 7921) described previously. Genomic DNA was prepared from the osteosarcoma patients samples by using a QIAamp DNeasy tissue kit (Qiagen, Valencia, CA) according to manufacturer instructions.
CGH. CGH was performed as described (32). Copy-number changes were detected based on the variance of the red/green ratio profile from the standard of 1. Ratio values of 1.2 and 2.0 were defined as thresholds for gains and high-level amplifications, respectively, and losses were defined as a ratio value of ≤0.8.
p53 Genotyping. Single-strand conformation polymorphism and DNA sequencing were performed essentially as described (33). Primers to amplify p53 exons 5–8 were purchased from Sigma Genosys (see Table 4, which is published as supporting information on the PNAS web site, www.pnas.org). DNA sequencing was performed by The Rockefeller University DNA Technology Center.
Multiplex PCR/ligase-detection reaction for p53, linked to read out on a universal DNA microarray, was performed as described (34). This technique allows for the detection of 110 specific p53 missense mutations in exons 5–8.
TaqMan Real-Time Quantitative PCR. HDM2 gene copy number was determined for each tumor relative to a control gene on chromosome 21, APP. Primers and TaqMan probes used for amplification and detection of both HDM2 and APP genes were purchased from Sigma Genosys (Table 4). PCR cycling conditions were 95°C for 10 min and 40 cycles of 95°C for 15 sec, 55°C for 30 sec, and 60°C for 30 sec, followed by one cycle of 60°C for 5 min. Reactions were performed by using the ABI 7700 sequence detector (TaqMan) from Applied Biosystems. Real-time quantitative assays were performed essentially as described (35). Briefly, standard curves were calculated for each gene by 10-fold serially diluting genomic DNA from normal placenta (Sigma). HDM2 gene copy numbers were determined by using the mean threshold cycle for each tumor, performed in triplicate, to calculate copy number from the HDM2 standard curve. APP control gene copy numbers were determined similarly from the APP standard curve and used for normalization of HDM2. Results are reported as HDM2 fold amplification relative to APP.
Statistical Tests. The correlation of p53 status with genomic-instability score was analyzed by using custom software written in matlab 6.5 (Mathworks, Natick, MA). Mock CGH data sets were generated under the null hypothesis of no correlation between p53 mutations and level of instability. Each mock data set was identical to the real set, i.e., contained the same number of tumors and changes per tumor but with random assignments of p53 mutant (n = 12) or HDM2-amplified (n = 5) tumor status. The mean number of chromosomal changes in p53 mutant and HDM2-amplified tumors in each such data set was calculated. The P value for the significance of the correlation of p53 status with genomic instability in the real data set was defined as the frequency of mock data sets that yielded a greater or equal mean number of changes per tumor than that of the real set (19.1) when 12 p53 mutant tumors were assigned randomly. The P value that expresses the significance of HDM2 amplification and its correlation to low-level instability was calculated similarly as the frequency of trials that yielded a mean number of changes that was lower or equal to that of the real set (8.8) when five HDM2-amplified tumors were assigned randomly. In both cases, 200,000 random data sets were generated.
Results
DNA Copy-Number Changes Identified by CGH. Genomic DNAs from 34 primary high-grade osteosarcoma tumors were subjected to CGH analysis to identify chromosomal copy-number changes relative to normal genomic DNA (Fig. 1 and Table 5, which is published as supporting information on the PNAS web site). Losses were detected on every autosome and the X chromosome in at least one tumor with the exception of chromosome 21. The most recurrent losses were of chromosomal regions 18q21q23 at 14/34 tumors (41%), followed by 6q16 at 13/34 tumors (38%) and 10q22q26 at 12/34 tumors (35%). Increases in copy number were detected on every autosome and the X chromosome in at least one tumor. The most frequently gained or amplified chromosomal regions were 17p11.2 and 6p12 at 16/34 (47%), followed by 1p32p36 at 15/34 (44%) and 8q24 at 15/34 (44%). Notably, 9 of the 16 tumors displaying increased copy number at 17p11.2, and 8 of 16 at 6p12, had high-level amplifications of these regions, which represent approximately ≥5-fold increases in copy number (36).
Fig. 1.
Ideogram of CGH-detectable copy-number changes in 34 osteosarcoma tumors. Copy-number gains are depicted by thin gray lines to the right of each chromosome, high-level amplifications (approximately >5-fold) are depicted by thick gray bars, and losses are depicted by thin black lines to the left of each chromosome.
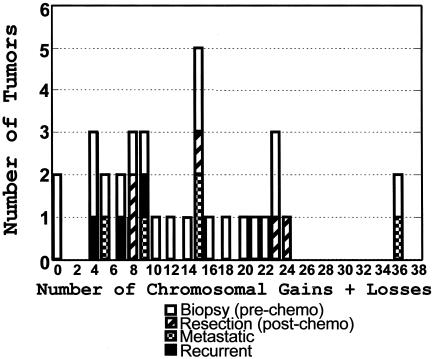
An instability score was assigned to each tumor, which is the sum of all losses, gains, and high-level amplifications detected in each sample (Fig. 2). Similar to a previous report of osteosarcoma CGH (30), a wide spectrum of genomic instability was found, with some tumors displaying few or no detectable changes and others displaying >20 chromosomal aberrations. The 34 tumors examined in this study include 21 tumor biopsies taken before chemotherapy, 5 resections taken after chemotherapy, 4 recurrent tumors from relapsed patients, and 4 metastatic tumors (Fig. 2).
Fig. 2.
Genomic-instability scores. The instability score for each sample is the total number of gains, losses, and high-level amplifications detectable by CGH. The CGH data from 34 high-grade osteosarcoma tumors (Fig. 1 and Table 5) are shown by sample type.
p53 Mutational Screening. Thirty-two of the 34 osteosarcomas were screened for mutations in the p53 DNA-binding domain by a combination of single-strand conformation polymorphism/sequencing and PCR/ligase-detection reaction/microarray techniques. The PCR/ligase-detection reaction technique is highly sensitive and allowed for the detection of mutations that otherwise were missed by single-strand conformation polymorphism (Table 1). p53 mutations were found in 12 of 32 tumors (38%),†† in exons 5–7, including three missense mutations at the commonly altered hotspot codons 175 and 273 (Table 1). Five other missense mutations occurred at codons 272, 281, 220, 224, and 173. Each of these mutations has been described in a variety of different cancers (37–42).
Table 1. p53 mutation screening.
| Tumor | p53 mutation | SSCP | PCR/LDR | |
|---|---|---|---|---|
| 1 | OS2 | Exon 8, D281H G → C | - | + |
| 2 | OS3 | Exon 8, V272M G → A | - | + |
| 3 | OS6 | Exon 5, R175H G → A | + | + |
| 4 | OS7 | Exon 6, frameshift | + | NT |
| 5 | OS8 | Exon 8, R273H G → A | - | + |
| 6 | OS11 | Exon 5, frameshift, del 17nt | + | NT |
| 7 | OS17 | Exon 6, E224D G → C | + | NT |
| 8 | OS19 | Exon 6, Y220C A → G | + | NT |
| 9 | OS22 | Exon 5, V173M G → A | + | NT |
| 10 | OS27 | Exon 5, deletion codon 190 | + | NT |
| 11 | OS28 | Exon 5, V173G T → G | + | NT |
| 12 | OS31 | Exon 8, R273C C → T | - | + |
SSCP, single-strand conformation polymorphism; LDR, ligase-detection reaction; +, found by technique; -, not found; NT, not tested; det 17nt, deletion of 17 nucleotides.
p53 mutant tumors were overrepresented among samples displaying high levels of genomic instability (Fig. 3a). The mean instability score for p53 mutant tumors was 19.1 chromosomal changes per tumor as compared with 11.1 for all other tumors. This correlation is statistically significant (P = 0.004) (Fig. 3b). The presence of mutant p53 was also significantly correlated with genomic instability in biopsy specimens (n = 21), with a mean of 19.9 changes in seven p53 mutant tumors versus 9.7 for all other biopsies (P = 0.009) (Fig. 4).
Fig. 3.
p53 mutation, HDM2 amplification, and genomic-instability score. (a) Osteosarcoma tumors with genomic amplification of HDM2 or mutation of p53 are depicted with corresponding genomic-instability scores. (b) P-value determination for p53 status and genomic-instability score. The probability distribution curve depicts the distribution of the mean numbers of genomic changes of 12 p53 mutant tumors of 200,000 randomized trials. The filled area underneath the curve (shown by the arrow) highlights those trials that produced a higher or equal mean number of changes to the actual mean (19.1).
Fig. 4.
Instability scores for osteosarcoma tumor biopsies. p53 mutant and HDM2-amplified osteosarcoma tumor biopsies are depicted with genomic-instability scores.
HDM2 Real-Time Quantitative PCR. Genomic amplification of the HDM2 gene was assayed by real-time quantitative PCR with TaqMan fluorescent probes. Gene copy numbers for HDM2 were normalized against the APP gene from chromosome 21. Amplification of this chromosome 21 locus has been shown to be extremely rare in a panel of breast carcinomas at a frequency of 0/108 (43). Also, in a previous CGH analysis of osteosarcomas, chromosome 21 was the least-commonly altered out of all chromosomes with one loss and two gains detected in total of 31 tumors (30). In the 34 osteosarcomas reported here, 5 of 34 osteosarcoma tumors were found to have ≥3-fold amplification of the HDM2 gene over the chromosome 21 control (Fig. 5). Three of these tumors had high-level amplifications of HDM2, from 11- to 21-fold.
Fig. 5.
Real-time quantitative PCR analysis of HDM2. The gene copy number for HDM2 for each tumor is shown relative to APP. Error bars represent SD.
In contrast to p53 mutant tumors, osteosarcomas displaying genomic amplification of HDM2 do not exhibit high levels of genomic instability (Figs. 3a and 4). The mean number of chromosomal aberrations for five HDM2-amplified tumors is 8.8. In this set of osteosarcomas, HDM2 oncogene amplifications actually cluster in the group of tumors with low levels of genomic instability (four of the five HDM2-amplified tumors are in the lower half of all tumors sorted by instability score) (Fig. 3a); however, this correlation is not significant (P = 0.1357).
Discussion
In this study, 34 high-grade osteosarcomas were analyzed for genome-wide chromosomal aberrations by CGH, and 32 of these were assayed for p53 mutations and genomic amplification of HDM2. The chromosomal abnormalities described in this set of tumors are similar in nature to a previous report with roughly the same number of high-grade osteosarcoma samples (n = 31) (30). A large number of genome alterations were detected, affecting every autosome and the X chromosome in at least one tumor, highlighting the breadth and magnitude of chromosomal instability in these tumors. There exists, above this genetic noise, a subset of genomic alterations that appear reproducibly in both studies (n = 65) (Tables 2 and 3). Copy-number increases at chromosomal regions 1q21, 8q24, 6p12, 8cenq13, 1p31p32, and 17p11.2 are the most significantly reproducible of 65 osteosarcomas. These regions of the osteosarcoma genome are likely to harbor genes that play a significant role in the initiation and/or propagation of tumorigenesis as evidenced by their repeated amplification in uncultured tumors. Notably, in the CGH data reported here, regions 17p11.2 and 6p12 seem to be highly significant by both quantitative (47% and 44%) and qualitative criteria, because each region is highly reproducible with a small common region of overlap that often presents as a high-level amplification (Fig. 1).
Table 2. CGH summary of 65 osteosarcomas.
| This report (34 tumors)
|
Previous report* (31 tumors)
|
||
|---|---|---|---|
| Locus | No. of tumors (%) | Locus | No. of tumors (%) |
| Most frequent copy-number increases | |||
| 6p12 | 16/34 (47), 8† | 1q21 | 18/31 (58), 1 |
| 17p11.2 | 16/34 (47), 9 | 8q21.3q22 | 16/31 (52), 3 |
| 1p32p36 | 15/34 (44) | 8cen-q13 | 14/31 (45), 2 |
| 8q24 | 15/34 (44), 2 | 8q23q24 | 13/31 (42), 1 |
| 20q | 11/34 (32), 1 | 14q24qter | 11/31 (35), 2 |
| Xp21 | 11/34 (32) | Xp11.2p21 | 11/31 (35), 4 |
| 8cen-q13 | 10/34 (29), 1 | 5p14 | 10/31 (32), 2 |
| 5p13-cen | 10/34 (29) | 6p12p21.3 | 10/31 (32), 3 |
| 1q21 | 10/34 (29), 2 | 11q14 | 10/31 (32) |
| 17q21q25 | 7/34 (21), 1 | 1p32q21 | 9/31 (29), 2 |
| Most frequent copy-number losses | |||
| 18q21q23 | 14/34 (41) | 6q16 | 10/31 (32) |
| 6q16 | 13/34 (38) | 6q21q22 | 10/31 (32) |
| 10q22q26 | 12/34 (35) | 10q23qter | 9/31 (29) |
| 13q21q31 | 11/34 (32) | 10p12pter | 9/31 (29) |
| 10p | 8/34 (24) | 13q21 | 8/31 (26) |
Previous data are from Tarkkanen et al. (30).
Bold numerals indicate the number of all copy-number increases that were high-level amplifications.
Table 3. Most frequent genomic changes in 65 tumors.
| Locus | No. of tumors (%) |
|---|---|
| +1q21 | 28/65 (43), 3 |
| +8q24 | 28/65 (43), 3 |
| +6p12 | 26/65 (40), 11 |
| +8cen-q13 | 24/65 (38), 3 |
| +1p32 | 24/65 (37), 2 |
| +17p11.2 | 23/65 (35), 9 |
| -6q16 | 23/65 (35) |
| +Xp21 | 22/65 (34), 4 |
| -10q23q26 | 21/65 (32) |
| +5p14 | 20/65 (31), 2 |
| -10p12pter | 17/65 (26) |
Bold numerals indicate the number of all copy-number increases that were high-level amplifications.
The significant correlation of p53 mutations with high levels of genomic instability in these osteosarcomas (Figs. 3 and 4) is consistent with experimental data obtained in cell culture (17, 18) and data from other uncultured human tumors in which p53 has been demonstrated to control the stability of the genome (22–24). The mutation of p53 therefore may be one major genetic factor contributing to the extremely high levels of genomic instability in some high-grade osteosarcomas. These data support the hypothesis that the loss of p53 function contributes to tumorigenesis by the destabilization of the genome.
The inhibition of homologous recombination by wild-type p53 has been reported by a number of groups and is one potential mechanism for control over the stability of the genome by p53 (44–48). Interestingly, a recent report examined the effectiveness of various p53 mutants at inhibiting homologous recombination. A p53 protein with a mutated Arg-273 residue was the most severely impaired for inhibition of homologous recombination, whereas a mutation disrupting Asp-281 performed more similarly to wild-type p53 (48). In the present study, the Asp-281 mutant tumor had an instability score of 8, whereas two tumors with mutations disrupting Arg-273 had approximately two to three times more instability with scores of 15 and 23, suggesting that the ability of specific mutations to inhibit homologous recombination may correlate with the genomic-instability scores determined by CGH.
Surprisingly, amplification of the HDM2 gene, which occurred mutually exclusive of p53 mutations, did not correlate with high levels of genomic instability in the osteosarcoma tumors reported here. In fact, p53 mutant tumors displayed a genomic-instability score that is more than twice that of the average HDM2-amplified tumor. The Hdm2 oncoprotein functions as a direct negative regulator of p53. Negative regulation by Hdm2 is achieved by the (i) direct binding and blocking of the p53 transactivation domain (25, 49–51), (ii) Hdm2-mediated shuttling of p53 from the nucleus to the cytoplasm (52, 53), and (iii) E3-ligase function of Hdm2, which targets p53 for ubiquitin-mediated proteasomal degradation (54). Overexpression of HDM2 by gene amplification has been observed in many different cancers and has been demonstrated to inhibit p53-dependent transactivation and apoptosis (26, 55). Amplification of HDM2 is therefore considered to effectively “knock out” or inactivate p53 protein function and, in so doing, may constitute the genetic equivalent to a p53 mutation. Consistent with this idea, the mutation of p53 and amplifications of HDM2 are normally mutually exclusive events in tumors with rare exception (27). Because p53 mutations and HDM2 amplifications were mutually exclusive genetic events, it is tempting to hypothesize that, although both events may achieve the inhibition of certain p53-mediated functions, on some level the amplification of HDM2 may not be equivalent to a p53 mutation with respect to genomic instability. Recently it was reported that the inactivation of p53 by stable short hairpin RNA (shRNA)-mediated silencing in mice mimics the p53 gene knockout phenotype and contributes to tumorigenesis in the Eμ-myc mouse lymphoma model (56). Interestingly, different p53 shRNA constructs contributed to tumorigenesis to varying degrees based on their differing abilities to knockdown levels of p53 protein. Although the loss of p53 by genetic knockout results in marked genomic instability in this model, none of the tumors that developed as a result of shRNA-mediated p53 knockdown, which retain residual levels of p53 protein, displayed high levels of genomic instability. These results and the data from human tumors presented here suggest that reducing the levels of wild-type p53 protein, endogenously by HDM2 amplification or exogenously by RNA interference, may contribute to tumorigenesis by inhibiting some p53 functions while still allowing for the maintenance of a relatively stable tumor genome by residual concentrations of p53. Further experiments are required to determine whether other human tumors with amplifications of HDM2 behave similarly and maintain relatively stable genomes as compared with tumors harboring a mutation in p53.
Supplementary Material
Acknowledgments
We thank members of the Levine Laboratory for helpful discussions, especially Drs. Kenan Onel, Archontoula Stoffel, Shengkan Jin, and Gareth Bond and Christine Walsh.
Abbreviation: CGH, comparative genome hybridization.
Footnotes
Several tumor samples were selected for this study based on the prior knowledge that they stained positively for p53 by immunohistochemistry (data not shown).
References
- 1.Hollstein, M., Sidransky, D., Vogelstein, B. & Harris, C. C. (1991) Science 253, 49–53. [DOI] [PubMed] [Google Scholar]
- 2.Levine, A. J., Momand, J. & Finlay, C. A. (1991) Nature 351, 453–456. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, W. P., Hollstein, M. C., Hsu, I. C., Sidransky, D., Lane, D. P., Vogelstein, B. & Harris, C. C. (1992) Chest 101, 19S–20S. [DOI] [PubMed] [Google Scholar]
- 4.Oliner, J. D., Kinzler, K. W., Meltzer, P. S., George, D. L. & Vogelstein, B. (1992) Nature 358, 80–83. [DOI] [PubMed] [Google Scholar]
- 5.Kamijo, T., Zindy, F., Roussel, M. F., Quelle, D. E., Downing, J. R., Ashmun, R. A., Grosveld, G. & Sherr, C. J. (1997) Cell 91, 649–659. [DOI] [PubMed] [Google Scholar]
- 6.Sherr, C. J. (2001) Nat. Rev. Mol. Cell Biol. 2, 731–737. [DOI] [PubMed] [Google Scholar]
- 7.Malkin, D. (1993) Cancer Genet. Cytogenet. 66, 83–92. [DOI] [PubMed] [Google Scholar]
- 8.Donehower, L. A., Harvey, M., Slagle, B. L., McArthur, M. J., Montgomery, C. A., Jr., Butel, J. S. & Bradley, A. (1992) Nature 356, 215–221. [DOI] [PubMed] [Google Scholar]
- 9.Tsukada, T., Tomooka, Y., Takai, S., Ueda, Y., Nishikawa, S., Yagi, T., Tokunaga, T., Takeda, N., Suda, Y., Abe, S., et al. (1993) Oncogene 8, 3313–3322. [PubMed] [Google Scholar]
- 10.Jacks, T., Shih, T. S., Schmitt, E. M., Bronson, R. T., Bernards, A. & Weinberg, R. A. (1994) Nat. Genet. 7, 353–361. [DOI] [PubMed] [Google Scholar]
- 11.Purdie, C. A., Harrison, D. J., Peter, A., Dobbie, L., White, S., Howie, S. E., Salter, D. M., Bird, C. C., Wyllie, A. H., Hooper, M. L., et al. (1994) Oncogene 9, 603–609. [PubMed] [Google Scholar]
- 12.Jin, S. & Levine, A. J. (2001) J. Cell Sci. 114, 4139–4140. [DOI] [PubMed] [Google Scholar]
- 13.Zhao, R., Gish, K., Murphy, M., Yin, Y., Notterman, D., Hoffman, W. H., Tom, E., Mack, D. H. & Levine, A. J. (2000) Genes Dev. 14, 981–993. [PMC free article] [PubMed] [Google Scholar]
- 14.Ko, L. J. & Prives, C. (1996) Genes Dev. 10, 1054–1072. [DOI] [PubMed] [Google Scholar]
- 15.Levine, A. J. (1997) Cell 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 16.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 17.Livingstone, L. R., White, A., Sprouse, J., Livanos, E., Jacks, T. & Tlsty, T. D. (1992) Cell 70, 923–935. [DOI] [PubMed] [Google Scholar]
- 18.Yin, Y., Tainsky, M. A., Bischoff, F. Z., Strong, L. C. & Wahl, G. M. (1992) Cell 70, 937–948. [DOI] [PubMed] [Google Scholar]
- 19.Burt, E. C., James, L. A., Greaves, M. J., Birch, J. M., Boyle, J. M. & Varley, J. M. (2000) Br. J. Cancer 83, 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukasawa, K., Wiener, F., Vande Woude, G. F. & Mai, S. (1997) Oncogene 15, 1295–1302. [DOI] [PubMed] [Google Scholar]
- 21.Venkatachalam, S., Shi, Y. P., Jones, S. N., Vogel, H., Bradley, A., Pinkel, D. & Donehower, L. A. (1998) EMBO J. 17, 4657–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgiades, I. B., Curtis, L. J., Morris, R. M., Bird, C. C. & Wyllie, A. H. (1999) Oncogene 18, 7933–7940. [DOI] [PubMed] [Google Scholar]
- 23.Eyfjord, J. E., Thorlacius, S., Steinarsdottir, M., Valgardsdottir, R., Ogmundsdottir, H. M. & Anamthawat-Jonsson, K. (1995) Cancer Res. 55, 646–651. [PubMed] [Google Scholar]
- 24.Primdahl, H., Wikman, F. P., von der Maase, H., Zhou, X. G., Wolf, H. & Orntoft, T. F. (2002) J. Natl. Cancer Inst. 94, 216–223. [DOI] [PubMed] [Google Scholar]
- 25.Momand, J., Zambetti, G. P., Olson, D. C., George, D. & Levine, A. J. (1992) Cell 69, 1237–1245. [DOI] [PubMed] [Google Scholar]
- 26.Chen, J., Wu, X., Lin, J. & Levine, A. J. (1996) Mol. Cell. Biol. 16, 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Momand, J., Jung, D., Wilczynski, S. & Niland, J. (1998) Nucleic Acids Res. 26, 3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spina, V., Montanari, N. & Romagnoli, R. (1998) Eur. J. Radiol. 27, Suppl. 1, S98–S109. [DOI] [PubMed] [Google Scholar]
- 29.Tarkkanen, M., Karhu, R., Kallioniemi, A., Elomaa, I., Kivioja, A. H., Nevalainen, J., Bohling, T., Karaharju, E., Hyytinen, E., Knuutila, S., et al. (1995) Cancer Res. 55, 1334–1338. [PubMed] [Google Scholar]
- 30.Tarkkanen, M., Elomaa, I., Blomqvist, C., Kivioja, A. H., Kellokumpu-Lehtinen, P., Bohling, T., Valle, J. & Knuutila, S. (1999) Int. J. Cancer 84, 114–121. [DOI] [PubMed] [Google Scholar]
- 31.Stock, C., Kager, L., Fink, F. M., Gadner, H. & Ambros, P. F. (2000) Genes Chromosomes Cancer 28, 329–336. [DOI] [PubMed] [Google Scholar]
- 32.Singh, B., Gogineni, S. K., Sacks, P. G., Shaha, A. R., Shah, J. P., Stoffel, A. & Rao, P. H. (2001) Cancer Res. 61, 4506–4513. [PubMed] [Google Scholar]
- 33.Suzuki, Y., Orita, M., Shiraishi, M., Hayashi, K. & Sekiya, T. (1990) Oncogene 5, 1037–1043. [PubMed] [Google Scholar]
- 34.Favis, R. & Barany, F. (2000) Ann. N.Y. Acad. Sci. 906, 39–43. [DOI] [PubMed] [Google Scholar]
- 35.Goff, L. K., Neat, M. J., Crawley, C. R., Jones, L., Jones, E., Lister, T. A. & Gupta, R. K. (2000) Br. J. Haematol. 111, 618–625. [DOI] [PubMed] [Google Scholar]
- 36.Weiss, M. M., Hermsen, M. A., Meijer, G. A., van Grieken, N. C., Baak, J. P., Kuipers, E. J. & van Diest, P. J. (1999) Mol. Pathol. 52, 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toguchida, J., Yamaguchi, T., Ritchie, B., Beauchamp, R. L., Dayton, S. H., Herrera, G. E., Yamamuro, T., Kotoura, Y., Sasaki, M. S., Little, J. B., et al. (1992) Cancer Res. 52, 6194–6199. [PubMed] [Google Scholar]
- 38.Foulkes, W. D., Stamp, G. W., Afzal, S., Lalani, N., McFarlane, C. P., Trowsdale, J. & Campbell, I. G. (1995) Br. J. Cancer 72, 883–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goh, H. S., Yao, J. & Smith, D. R. (1995) Cancer Res. 55, 5217–5221. [PubMed] [Google Scholar]
- 40.Kannio, A., Ridanpaa, M., Koskinen, H., Partanen, T., Anttila, S., Collan, Y., Hietanen, E., Vainio, H. & Husgafvel-Pursiainen, K. (1996) Cancer Epidemiol. Biomarkers Prev. 5, 33–39. [PubMed] [Google Scholar]
- 41.Ostwald, C., Gogacz, P., Hillmann, T., Schweder, J., Gundlach, K., Kundt, G. & Barten, M. (2000) Int. J. Cancer 88, 82–86. [DOI] [PubMed] [Google Scholar]
- 42.Schlechte, H. H., Sachs, M. D., Lenk, S. V., Brenner, S., Rudolph, B. D. & Loening, S. A. (2000) Cancer Detect. Prev. 24, 24–32. [PubMed] [Google Scholar]
- 43.Bieche, I., Olivi, M., Champeme, M. H., Vidaud, D., Lidereau, R. & Vidaud, M. (1998) Int. J. Cancer 78, 661–666. [DOI] [PubMed] [Google Scholar]
- 44.Dudenhoffer, C., Kurth, M., Janus, F., Deppert, W. & Wiesmuller, L. (1999) Oncogene 18, 5773–5784. [DOI] [PubMed] [Google Scholar]
- 45.Gebow, D., Miselis, N. & Liber, H. L. (2000) Mol. Cell. Biol. 20, 4028–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willers, H., McCarthy, E. E., Wu, B., Wunsch, H., Tang, W., Taghian, D. G., Xia, F. & Powell, S. N. (2000) Oncogene 19, 632–639. [DOI] [PubMed] [Google Scholar]
- 47.Willers, H., McCarthy, E. E., Alberti, W., Dahm-Daphi, J. & Powell, S. N. (2000) Int. J. Radiat. Biol. 76, 1055–1062. [DOI] [PubMed] [Google Scholar]
- 48.Akyuz, N., Boehden, G. S., Susse, S., Rimek, A., Preuss, U., Scheidtmann, K. H. & Wiesmuller, L. (2002) Mol. Cell. Biol. 22, 6306–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliner, J. D., Pietenpol, J. A., Thiagalingam, S., Gyuris, J., Kinzler, K. W. & Vogelstein, B. (1993) Nature 362, 857–860. [DOI] [PubMed] [Google Scholar]
- 50.Chen, J., Lin, J. & Levine, A. J. (1995) Mol. Med. 1, 142–152. [PMC free article] [PubMed] [Google Scholar]
- 51.Kussie, P. H., Gorina, S., Marechal, V., Elenbaas, B., Moreau, J., Levine, A. J. & Pavletich, N. P. (1996) Science 274, 948–953. [DOI] [PubMed] [Google Scholar]
- 52.Roth, J., Dobbelstein, M., Freedman, D. A., Shenk, T. & Levine, A. J. (1998) EMBO J. 17, 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao, W. & Levine, A. J. (1999) Proc. Natl. Acad. Sci. USA 96, 3077–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang, S., Jensen, J. P., Ludwig, R. L., Vousden, K. H. & Weissman, A. M. (2000) J. Biol. Chem. 275, 8945–8951. [DOI] [PubMed] [Google Scholar]
- 55.Haupt, Y., Barak, Y. & Oren, M. (1996) EMBO J. 15, 1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 56.Hemann, M. T., Fridman, J. S., Zilfou, J. T., Hernando, E., Paddison, P. J., Cordon-Cardo, C., Hannon, G. J. & Lowe, S. W. (2003) Nat. Genet. 33, 396–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.