Abstract
MHC typing for human hematopoietic cell transplantation (HCT) from unrelated donors is currently performed by using a combination of serologic and molecular techniques. It has been determined that allelic differences in human MHC molecules, revealed by nucleotide sequencing but not by serologic typing, substantially influence graft rejection and graft-versus-host disease, two serious complications of clinical HCT. We studied transplantation of purified hematopoietic stem cells in a series of mouse strains that were matched at the MHC but had different background genes, and we observed striking differences in engraftment resistance and graft-versus-host disease severity, both factors depending on the donor–recipient strain combination. The individual mouse lines studied here were established nearly a century ago, and their MHC types were determined exclusively by serologic techniques. We considered the possibility that serologically silent MHC polymorphisms could account for our observations and, therefore, we performed DNA sequencing of the class I and II MHC alleles of our mouse strains. At each locus, exact homology was found between serologically MHC-matched strains. Our results likely extend to all serologically MHC-matched mouse strains used in modern research and highlight the profound and variable influence that non-MHC genetic determinants can have in dictating outcome after HCT.
Keywords: histocompatibility antigens, hematopoietic stem cell transplantation
In clinical allogeneic hematopoietic cell transplantation (HCT), matching of donors and recipients at the MHC is considered essential to minimize graft-versus-host disease (GVHD) and graft rejection. Related donors are usually siblings who have inherited the same parental alleles and thus encode MHC molecules (designated HLA in humans) genotypically identical to the patient. Because related donors are often unavailable, MHC-matched unrelated donors are sought by volunteer bone marrow (BM) donor registries. Historically, human MHC typing has been performed by using panels of alloantibodies derived from individuals exposed to HLAs by pregnancy or transfusion (1). In the last decade, such serologic typing has given way to the more precise DNA-based technology, which has revealed that in >20% of serologically matched unrelated donors, sequence polymorphisms exist in class I and/or class II MHC molecules (2–7). Thus, serologically defined HLAs can encode families of alleles whose differences are distinguishable only by DNA-based typing methods. These polymorphisms encode amino acid substitutions that cluster within the MHC peptide binding groove and, although serologically silent (2, 8), can influence T cell activation. It has been shown in human HCT that the consequences of these serologically undetectable differences include increased risks of GVHD and mortality when the disparities are in the MHC class II loci (7, 9) and increased graft failures in the cases of MHC class I disparities (2, 3, 10).
Polymorphisms at non-MHC genes encoding for minor histocompatibility antigens (mHAgs) can also induce GVHD and/or graft rejections. mHAgs are peptide antigens derived from normal cellular proteins that elicit MHC-restricted T cell responses (11). The importance of mHAgs in human transplantation is proven by the observation that even MHC-identical sibling pairs can develop these complications, although the frequency and degree of complications are highly variable among donor–recipient pairs. The identity of the mHAg(s) that consistently cause these complications has not been determined. Both quantitative and qualitative genetic differences between donor and recipient have been proposed to explain the observed heterogeneities. A simple quantitative explanation is that the number of mismatched mHAgs differs among different donor–recipient pairs. Qualitative explanations include the hypothesis that the immunogenicity of different mHAgs may vary depending on the pool of responding T cells selected according to the unique set of background genes in the donor versus recipient, or alternatively, on polymorphisms of immune regulatory molecules, which dampen or augment immune responses (12) and interact with mismatched mHAgs to modify GVHD and other complications (13, 14).
To better understand and characterize these non-MHC genetic factors of allogeneic HCT in an experimental model, we studied transplantation of purified hematopoietic stem cells (HSC) in a series of mouse strains that were matched at the MHC but had different background genes. It has been demonstrated by us (15) and others (16–18) that unfractionated BM contains non-HSC populations that can facilitate HSC engraftment. When HSC alone are transplanted, they are more strongly resisted as compared with BM. Thus, transplantation of HSC alone allowed us to detect the effects of subtle differences mediated by mHAgs in engraftment resistance in the absence of a confounding graft-facilitating population.
We observed striking differences in engraftment resistance and GVHD severity, both of which depended on the donor–recipient strain combination. The mouse strains used in this study diverged more than a century ago (Fig. 1), and their MHC typing has been determined by serologic methods. Given the role of MHC sequence differences in determining outcomes after allogeneic HCT and the highly polymorphic nature of MHC molecules, we considered that the dramatic differences in outcome observed in our mouse models might have been influenced by previously unrecognized polymorphisms in the mouse MHC, rather than by non-MHC determinants alone. Therefore, we sequenced the MHC class I and II loci of the five H2b or H2k mouse strains used in our studies and compared the outcome with available nucleotide sequences for the mouse H2b or H2k haplotypes from public sequence databases.
Fig. 1.
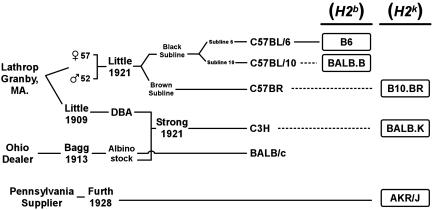
Genealogy of inbred mice used in this study. Aligned horizontally are the MHC donors for the congenic strains BALB.B, B10.BR, and BALB.K, which were C57BL/10J, C57BR/cdJ, and C3H/He mice, respectively, as indicated by dashed lines. B6 is the Thy1 C57BL/6 congenic strain expressing the Thy1.1 allele used as HSC donors for BALB.B mice in transplant experiments. The non-MHC background genes were of diverse origin as described in the text.
Three MHC-matched strain combinations with diverse origins were studied: one H2b (B6 → BALB.B) and two H2k (AKR/J → B10.BR and AKR/J → BALB.K) matched. These five strains belong to distinct lineages of the laboratory mouse, and their genealogical derivation is shown in Fig. 1. The AKR/J, BALB/c, and combined C57BL/6 and C57BL/10 sublines were independently derived founding lines and provide nonoverlapping background genes for each pair (19). The MHC donor for BALB.B was C57BL/10, which is closely related to C57BL/6 because both strains originally cosegregated in the black subline from crosses performed by C. C. Little (The Jackson Laboratory) with male and female littermates provided by Abby Lathrop (Granby, MA) (20). In contrast, the MHC of the H2k mice in this study was derived from three separate origins. C3H and C57BR mice were MHC donors for the BALB.K and B10.BR MHC congenic lines, respectively. The C3H strains were developed by Strong from a cross of a Bagg albino female with a DBA stock and then selected for a high incidence of mammary tumors (21). C57BR was inbred from the brown subline of the same cross that gave rise to founders for the C57BL strains. AKR/J was established by Furth from a leukemia-prone line that shared no clear common lineage to either C3H or C57BR (22).
Materials and Methods
Mice. All mice were bred and maintained at the Stanford University Research Animal Facility. Thy1.1 congenic C57BL/Ka (H2b, Thy1.1) and AKR/J(H2k, Thy1.1) mice served as donor strains in HSC transplantation experiments and were 6–10 weeks of age. The C57BL/Ka line was derived from C57BL/6 mice introduced into our colony at Stanford University by Henry Kaplan more than 30 years ago and will be referred to as B6 in the text. MHC congenic BALB.B (H2b, Thy1.2), B10.BR (H2k, Thy1.2), and BALB.K (H2k, Thy1.2) recipient mice were >8 weeks of age at the time of lethal irradiation.
HSC Transplantation. Adult HSC were isolated by a modified procedure described by Spangrude et al. (23). In brief, freshly harvested BM cells were selected for c-Kit (3C11) positivity with MidiMACS separation units (Miltenyi Biotec, Auburn, CA) and then sorted with multiparameter fluorescence-activated cell sorting (FACS) for a c-Kit+ (2B8), Thy1.1lo-int (19XE5), and Sca-1+ (E13-161) population that lacked expression of lineage markers (CD3, 145-2C11; CD4, GK1.5; CD5, 53-7.8; CD8, 53-6.7; B220, 6B2; Gr1, 8C5; Mac1, M1/70; and Ter119). Purified HSC were infused by lateral tail-vein injection into recipient mice within 24 h of irradiation. BALB.B and BALB.K mice were given 800 cGy, and B10.BR mice were given 950 cGy of whole-body γ irradiation, delivered in two split doses >4 h apart. Survival after HSC transplantation was monitored daily for >100 days after transplantation.
GVHD Induction and Analysis. Single-cell suspensions of unseparated whole splenocytes were coinjected with 3,000 HSC from donor mice into lethally irradiated recipient mice. A dose of 3,000 purified HSC is fully radioprotective for all three MHC-matched mouse strain combinations. Whole splenocyte doses were 1.0 × 107,1.5 × 107, and 2.0 × 107 for BALB.K, B10.BR, and BALB.B mice, respectively. Survival, weight loss, and clinical signs of GVHD were monitored daily for 60 days after transplantation. Liver, terminal ileum, and skin were obtained at day 60 from surviving mice for histopathological analysis by an expert pathologist who was without knowledge of the experimental group. The GVHD scoring system has been described (24).
Analysis of Engraftment by FACS and PCR. Hematopoietic chimerism in the T cell compartment was assessed by FACS analysis using monoclonal antibodies against Thy1.1 (Ox-7) expressed by donor mice and Thy1.2 (53-2.1) expressed by recipient mice. Donor chimerism in the B cell and granulocyte compartments was determined by PCR for microsatellite marker targets on genomic DNA isolated from FACS-sorted B220+ and Mac1/Gr1+ peripheral blood cells by using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Marker D8Mit224 provided informative polymorphisms for B6 → BALB.B and AKR/J → B10.BR transplants, whereas marker D6Mit3 was used for AKR → BALB.K recipients (25). PCR was run with 100 ng of genomic DNA and 20 pmol of each primer in a 50-μl reaction volume containing 1.5 mM MgCl2 for 35 cycles with an annealing temperature of 57°C. Ethidium bromide-stained PCR products were analyzed by agarose gel electrophoresis.
MHC Class I and II Genotyping. For each mouse strain, cDNA sequencing readings through all exons encoding for external protein domains at the H2-K, H2-D, H2-E, and H2-A loci were obtained by using methodologies similar to those used for high-resolution MHC allele typing in humans (26). Total RNA was purified from unstimulated whole splenocytes harvested from retired breeders with the RNeasy Mini Kit (Qiagen) and reverse transcribed to cDNA by using oligo(dT) primers and SuperScript II (Life Technologies, Grand Island, NY) according to the manufacturer's instructions. PCR was performed with locus-specific primers for 20–30 cycles by using 2-μl aliquots of the cDNA template and an annealing temperature optimized for each primer set. PCR amplicons were purified with ExoSAP-IT (United States Biochemical) and sequenced by the DYEnamic ET dye terminator cycle sequencing reaction (Amersham Biosciences) in both forward and reverse directions by using a second set of locus-specific sequencing primers.
MHC Locus-Specific PCR Primers. H2K and H2D are mouse class I MHC antigens that form a complex with β2-microglobulin on the cell surface. H2-A and H2-E α and β heterodimers are the class II MHC antigens expressed in our mouse strains. PCR primers for the MHC class I H2-K and H2-D loci amplified exons 2–4. PCR primers for the MHC class II H2-Aα, H2-Aβ, H2-Eα, and H2-Eβ loci amplified exons 2 and 3. Primer sequences were as follows: H2-K, 5′-GGAACTCAGAAGTCTCGAATCG-3′ and 5′-CCACAGCTCCAGTGACTATTGC-3′; H2-D, 5′-ACACCCGGGATCCCAGATG-3′ and 5′-CACAGCTCCAATGATGGCCA-3′; H2-Aα, 5′-GACCTCCCGGAGACCAGG-3′ and 5′-CGCCTTCCTTTCCAGGGT-3′; H2-Aβ,5′-CCTCTTGGCTGCTGTTGTG-3′ and 5′-TCCTTTCTGACTCCTGTGACG-3′; H2-Eα, 5′-ATTGGAGCCCTGGTGTTA-3′ and 5′-TCCTTGTCGGCGTTCTAC-3′; and H2-Eβ, 5′-CTGTGTGGCAGCTGTGATCCT-3′ and 5′-CCTGTTGGCTGAAGTCCAGACT-3′.
MHC Locus-Specific Sequencing Primers. A second set of oligonucleotides nested internally to PCR primers for each MHC locus was used for the sequencing reaction. Forward and reverse sequencing primer sequences, respectively, were as follows: H2-K, 5′-CTGCATGCTGCTCCTGCTGT-3′ and 5′-AAGGACAACCAGAACAGCAA-3′; H2-D, 5′-GCACGCTGCTCCTGCTGC-3′ and 5′-AGGACACCCAGAACAGCAA-3′; H2-Aα, 5′-GTCCTCGCCCTGACCACC-3′ and 5′-ACCACGATGCCCACAAGG-3′; H2-Aβ, 5′-CAGCCCAGGGACTGAGGG-3′ and 5′-GCCAAGCCCGAGGAAGAT-3′; H2-Eα, 5′-CCAGAAGTCATGGGCTATC-3′ and 5′-AGACCCACAAACAACCCAAGAG-3′; and H2-Eβ, 5′-GCTGACAGTGCTGAGCCCTC-3′ and 5′-AGCACGAAGCCCCCAACT-3′.
GenBank Accession Numbers. GenBank mRNA sequences and their accession numbers for MHC class I and II loci used as reference sequences for alignment are as follows: H2-Kb, U47328; H2-Kk, U47330; H2-Db, U47325; H2-Dk, U47327; H2-Aαb, K01922; H2-Aαk, V00832; H2-Aβk, M13538; H2-Eβb, AK012147; and H2-Eβk, K01145.
Statistical Analysis. Survival was analyzed by using the Kaplan–Meier method, and differences among experimental groups were assessed by using the log rank test. Differences in the median observations were determined by using the Wilcoxon rank sum test. Statistical significance was conferred at P < 0.05.
Results
Quantitation of Resistance to Engraftment. In three MHC-matched mouse strain combinations (B6 → BALB.B, AKR/J → B10.BR, and AKR/J → BALB.K) resistance to allogeneic hematopoietic cell engraftment and GVHD was compared by transplanting purified HSC or HSC plus splenocytes, respectively. Engraftment resistance can be quantitated as the minimal HSC dose required for rescue of irradiated recipients (15). In allogeneic HSC transplantation, engraftment resistance correlates with increased genetic disparity, with the strongest barriers observed in MHC mismatched mice. Furthermore, purified allogeneic HSC encounter greater resistance to engraftment as compared with unfractionated BM because the latter contains non-HSC populations that facilitate engraftment (16). Even in highly disparate genetic combinations, resistance generally can be overcome by administration of high HSC doses. Host-versus-graft immune responses between MHC-matched mice, on the other hand, may be subtle and may require the accurate and reliable quantitation of graft rejection by an HSC radioprotection assay to detect.
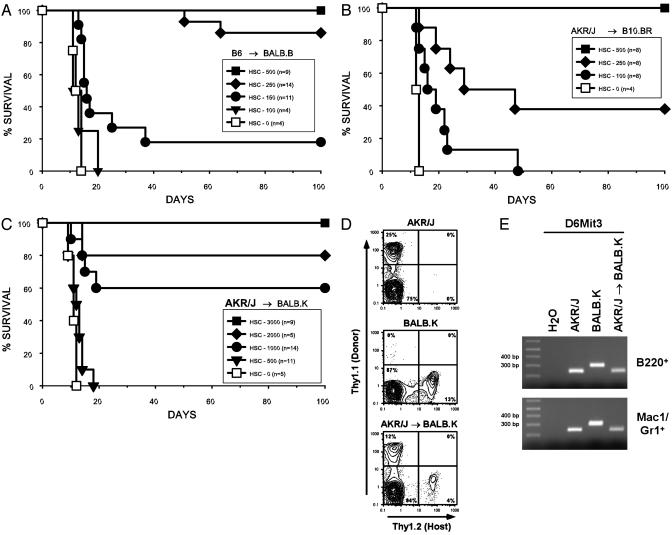
Purified HSC were isolated from mouse BM by FACS based on a composite surface phenotype of high staining for c-Kit and Sca-1, intermediate staining for Thy-1.1, and negative staining for lineage-specific markers. The Thy-1loLin–Sca-1+c-Kit+ cells constitute a rare population in mouse BM and are enriched 2,000-fold for HSC activity. One hundred to two hundred of such HSC-enriched cells consistently rescue 90–100% of lethally irradiated syngeneic recipient mice (27), whereas ≈1–2 × 105 unfractionated syngeneic BM cells are required to achieve equivalent radioprotection (28). Titrated numbers of purified HSC were injected after lethal irradiation of recipients. In the B6 → BALB.B combination, the barrier to engraftment was essentially at the same low level required for radioprotection of syngeneic/congenic recipients (23, 27), with only 250 HSC rescuing >80% and 500 HSC rescuing 100% of irradiated recipients (Fig. 2A). The HSC dose required for radioprotection was slightly higher (P = 0.01) in AKR/J → B10.BR transplants, in which 250 HSC rescued <40% but 500 HSC still rescued 100% of recipient mice (Fig. 2B). The highest (P < 0.001) HSC rescue dose requirement was observed in the AKR/J → BALB.K combination, in which 500 HSC rescued 0% and >3,000 HSC were required to rescue 100% of recipients (Fig. 2C). The 3,000 HSC dose needed for AKR/J rescue of irradiated BALB.K mice approaches the HSC dose requirement for rescue of MHC-mismatched mice (15) and represents a >6-fold increase compared with the B6 → BALB.B and AKR/J → B10.BR transplants. Multilineage donor engraftment was confirmed >6 weeks posttransplantation in surviving mice by FACS analysis for T cell chimerism (Fig. 2D) and in some instances by PCR testing of genomic DNA extracted from B220+ and Mac1+/Gr1+ blood cells sorted by FACS for microsatellite markers informative among donor–recipient pairs (Fig. 2E). All surviving mice were reconstituted with full B cell, granulocyte, and mixed T cell donor chimerism, which is characteristic of irradiated mice rescued with purified HSC (15, 16, 29).
Fig. 2.
Variable resistance to HSC engraftment in MHC-matched mice. BALB.B, B10.BR, and BALB.K mice were lethally irradiated and given titrated doses of Thy-1loLin–Sca-1+c-Kit+ HSC from MHC-matched donors. Strain combinations transplanted were B6 → BALB.B (A), AKR/J → B10.BR (B), and AKR/J → BALB.K (C) recipients. BALB.B and BALB.K mice were irradiated with 800 cGy, whereas B10.BR mice received 950 cGy. Results are pooled from at least two experiments for each mouse pair. Also shown are representative peripheral blood hematopoietic chimerism studies in AKR/J → BALB.K mice 60 days after irradiation and transplantation with 3,000 HSC demonstrating donor T cell engraftment by FACS (D) and B cell and granulocyte engraftment by PCR using microsatellite marker D6Mit3 (E). PCR amplicon sizes at D6Mit3 are 239 bp for AKR/J and 300 bp for BALB.K mice.
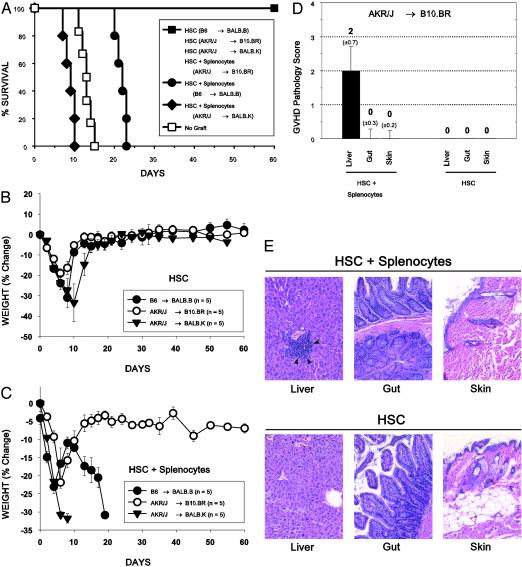
GVHD. Severity of GVHD in these MHC-matched mouse strains was determined by studying survival (Fig. 3A) and weight change (Fig. 3 B and C) after cotransplantation of 1.0–2.0 × 107 splenocytes plus purified HSC into lethally irradiated mice. All control irradiated mice given MHC-matched HSC alone survived to day 60 without evidence of GVHD and recovered to baseline weight, which was maintained until the end of the experiment. Irradiated BALB.B mice given B6 HSC plus splenocytes died at a median of 22 days with progressive weight loss and clinical signs of acute GVHD, including hunched posture and diarrhea. This severity of GVHD was consistent with prior observations involving the B6 → BALB.B strain combination (30). Somewhat more striking, BALB.K mice given AKR/J splenocytes plus HSC developed an aggressively lethal GVHD with a median survival time of only 9 days. In contrast, B10.BR mice given AKR/J HSC with splenocytes developed only clinically mild GVHD (primarily manifested by a failure to recover beyond 5% below baseline weight) without lethality by day 60. The mild GVHD in surviving B10.BR recipients of AKR/J HSC plus splenocytes was further studied by histological examination of tissue at 60 days posttransplantation. Skin, terminal ileum, and liver, the tissue targets of acute GVHD, revealed histologic features consistent with mild GVHD limited to the liver (Fig. 3 D and E). No significant evidence of GVHD was noted in the other tissues examined or in the tissues of recipients of HSC only.
Fig. 3.
Variation of GVHD severity in MHC-matched mice. (A–C) Survival and percent weight change after lethal irradiation and transplantation of 3,000 Thy-1loLin–Sca-1+c-Kit+ HSC, alone or with whole splenocytes. Recipients of purified HSC grafts did not develop signs of GVHD. B10.BR recipients of AKR/J HSC plus splenocytes recovered to within 5% of baseline weight but did not display overt signs of GVHD, and all mice survived to day 60. In contrast, transplantation of HSC plus splenocytes into irradiated BALB.B and BALB.K mice from B6 and AKR/J donors, respectively, resulted in lethal GVHD that was particularly aggressive in the latter, with a median survival time of 9 days (P = 0.003, compared with B10.BR mice given AKR/J HSC plus splenocytes). (D and E) Liver, terminal ileum, and skin were obtained from surviving B10.BR mice on day 60 for pathology assessment. Significant pathologic evidence of GVHD with a median score of 2 was limited to liver of B10.BR mice given HSC plus splenocytes (P = 0.008, compared with mice given HSC alone), the histology of which revealed portal lymphoid infiltrates and focal bile duct invasion (arrowheads in E) without biliary ductal regeneration.
MHC Allele Typing by DNA Sequencing. The above experiments demonstrated wide variability in engraftment resistance and GVHD severity after HSC transplantation in three MHC-matched mouse pairs that recapitulate results seen with human HCT. Routine MHC typing in mice has been based solely on serologic characterization of MHC antigens with polyclonal antisera and monoclonal antibodies (31). In humans, however, it is known that a single serologically defined class I or II MHC antigen can contain an entire family of different alleles (32). These MHC molecular differences correlate with graft rejection and hold significant predictive value for severe GVHD (2–5, 9). DNA sequences of mouse class I and II MHC genes in public databases have come primarily from studies of structural diversity among different H2 alleles, and thus serologically defined alleles have rarely been sequenced in more than a single mouse strain (33). We considered, therefore, that a likely explanation for the observed differences among the mouse strain combinations tested here might be subtle MHC nucleotide sequence polymorphisms undetected by serologic analysis. To exclude the presence of such polymorphisms, nucleotide sequencing of H2-K, H2-D, H2-A, and H2-E cDNA was performed on all mouse strains in our study. As shown in Table 1, complete nucleotide sequence homology was observed at all expressed MHC class I and II loci when aligned for interstrain comparisons among the three MHC-matched mouse pairs. Furthermore, for each locus our sequences were in complete agreement with publicly accessible mRNA sequences matched for either H2b or H2k haplotypes. H2-Eα is transcriptionally silent in H2b mice (34) and, before this study, had not been sequenced for any H2k haplotype mouse strain. H2-Aβb genomic DNA has been sequenced for B6, and the predicted mRNA sequence matches our H2-Aβb cDNA sequence (35). We conclude, therefore, that complete nucleotide homology exists at the MHC matched among the strains in this study, and thus, serologically silent MHC mismatches are excluded as a possible explanation for our hematopoietic graft outcomes.
Table 1. Genotypic identity at MHC class I and II loci by cDNA sequencing.
| % nucleotide sequence homology
|
||||||
|---|---|---|---|---|---|---|
| MHC class I loci
|
MHC class II loci
|
|||||
| Strain | H2-K | H2-D | H2-Aα | H2-Aβ | H2-Eα | H2-Eβ |
| H2b haplotype | ||||||
| GenBank strain | C57BL/10 | C57BL/10 | 129 | NA | NA | B6 |
| B6 | 100 | 100 | 100 | 100 | — | 100 |
| BALB.B | 100 | 100 | 100 | 100 | — | 100 |
| H2k haplotype | ||||||
| GenBank strain | C3H | C3H | A/J | B10.A | NA | B10.A |
| AKR/J | 100 | 100 | 100 | 100 | 100 | 100 |
| B10.BR | 100 | 100 | 100 | 100 | 100 | 100 |
| BALB.K | 100 | 100 | 100 | 100 | 100 | 100 |
MHC locus-specific PCR amplicons from cDNA templates spanning exons 2-4 (class I genes) and exons 2-3 (class II genes) were directly sequenced in both forward and reverse directions. Shown is percent nucleotide sequence homology for each mouse strain with a GenBank sequence at each locus. Reference cDNA sequences were not available for H2-Aβb, H2-Eαb, and H2-Eαk; H2-Eα is not expressed in H2b haplotype strains. Percent homology at the H2-Aβb and H2-Eαk loci represents interstrain comparison of mice used in this study. NA, not available.
Discussion
Herein, we show significant variation in resistance to allogeneic hematopoietic cell engraftment and in development of GVHD for different donor/host mouse strain combinations that are MHC matched. Because the mice in our study carry nonoverlapping sets of background genes, these data highlight the role of non-MHC genetic determinants in influencing the variation in outcome after HCT. In addition, resistance to engraftment and GVHD appear to be controlled by separate determinants because in at least one of three strain combinations studied there was no correlation between resistance and GVHD development.
The cumulative effect of the mHAg differences among AKR/J → BALB.K recipients resulted in HSC engraftment resistance and GVHD severity proportionate with that expected from fully allogeneic mismatched mice disparate at major rather than minor histocompatibility loci. In contrast, HSC transplanted into BALB.B mice encountered little engraftment resistance, but transfer of B6 splenocytes plus HSC resulted in fulminant GVHD. The identity and function of the mHAgs that control these clinical outcomes have not been determined. For GVHD in the B6 → BALB.B strain combination, a strongly immunogenic mHAg (designated H60) that is encoded by a gene expressed on the BALB but not the C57BL/6 background has recently been described (36, 37). Although it is possible that polymorphisms in expressed gene products that serve as direct antigenic stimuli for alloreactive T cells are responsible for inducing responses in the host-versus-graft (resistance) or GVHD direction, more complex interactions may mediate these effects. These interactions can include epigenetic interplay between a mismatched mHAg and modifying loci, such as molecules that regulate immune responses that result in augmentation or attenuation of alloreactive responses. To understand more precisely the genetic basis of outcome after MHC-identical allogeneic HCT, it may be possible to identify genetic susceptibility loci in the BALB.K recipients of AKR grafts that induce or protect from GVHD by using a positional cloning strategy.
To study the impact of mHAgs here it was essential to formally exclude serologically silent MHC polymorphisms among different strains of inbred mice bearing the same serological H2 haplotype. The absence of such allelic subgroups within a serologically defined MHC haplotype had never been disproved and was considered a possibility because studies of wild mouse populations have shown H2 diversity to be rather extensive (38). Furthermore, exhaustive large-scale screening for rejection of parental tail-skin graft in a single mouse colony identified many intra-MHC mutations (H2bm) that arose spontaneously during the 25-year observation period (39). It is intriguing that these mutations, which occurred on class I as well as class II H2 antigens, often mirrored HLA polymorphisms by encoding for subtle amino acid substitutions at key contact residues in the peptide binding groove. We found no evidence for allelic subgroups in the haplotypes and strains in these experiments.
The fact that MHC polymorphisms were not present in the mouse strain combinations studied here further suggests that undetected polymorphisms are likely not present in most commonly used inbred mice that are serologically MHC identical. Collectively, the origins of the MHC genes in our mice trace back to DBA, BALB/c, both black and brown sublines of C57, and the independent AKR/J line. Although limited to H2b and H2k haplotypes, the mouse strains in this study reflect diverse MHC derivations that sample a significant cross section of major genealogical lineages (19, 40). Our findings likely reflect a population bottleneck derived from the limited number of often historically murky mouse suppliers (some now known only as “the dealer from Ohio” or “the Pennsylvania supplier”) who contributed to the founding lines of contemporary inbred mice nearly a century ago (19, 40). It is also possible that protection from parasites and microbial pathogens, due to housing in closed colonies, may have mitigated selective pressures that otherwise could have promoted adaptive MHC polymorphic evolution in modern inbred laboratory mice. Finally, the results of our studies are a testament to the care and accuracy of the mouse handlers who have bred these animals through many decades and avoided interstrain contaminations that could have resulted in intra-MHC recombinants.
Acknowledgments
We thank Peter Parham for thoughtful discussions and critical reading of the manuscript and Lucino Hidalgo for excellent care of our mouse colony. This work was supported in part by National Institutes of Health/National Cancer Institute Grant 2PO1CA049605.
Abbreviations: BM, bone marrow; FACS, fluorescence-activated cell sorting; GVHD, graft-versus-host disease; HCT, hematopoietic cell transplantation; HSC, hematopoietic stem cells; mHAg, minor histocompatibility antigen.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY303781–AY303785).
References
- 1.Mickelson, E. & Petersdorf, E. W. (1999) in Hematopoietic Cell Transplantation, eds. Thomas, E. D., Blume, K. G. & Forman, S. J. (Blackwell Scientific, Malden, MA), pp. 28–37.
- 2.Petersdorf, E. W., Hansen, J. A., Martin, P. J., Woolfrey, A., Malkki, M., Gooley, T., Storer, B., Mickelson, E., Smith, A. & Anasetti, C. (2001) N. Engl. J. Med. 345, 1794–1800. [DOI] [PubMed] [Google Scholar]
- 3.Morishima, Y., Sasazuki, T., Inoko, H., Juji, T., Akaza, T., Yamamoto, K., Ishikawa, Y., Kato, S., Sao, H., Sakamaki, H., et al. (2002) Blood 99, 4200–4206. [DOI] [PubMed] [Google Scholar]
- 4.Sasazuki, T., Juji, T., Morishima, Y., Kinukawa, N., Kashiwabara, H., Inoko, H., Yoshida, T., Kimura, A., Akaza, T., Kamikawaji, N., et al. (1998) N. Engl. J. Med. 339, 1177–1185. [DOI] [PubMed] [Google Scholar]
- 5.Petersdorf, E. W., Gooley, T. A., Anasetti, C., Martin, P. J., Smith, A. G., Mickelson, E. M., Woolfrey, A. E. & Hansen, J. A. (1998) Blood 92, 3515–3520. [PubMed] [Google Scholar]
- 6.Scott, I., O'Shea, J., Bunce, M., Tiercy, J. M., Arguello, J. R., Firman, H., Goldman, J., Prentice, H. G., Little, A. M. & Madrigal, J. A. (1998) Blood 92, 4864–4871. [PubMed] [Google Scholar]
- 7.Speiser, D. E., Tiercy, J. M., Rufer, N., Grundschober, C., Gratwohl, A., Chapuis, B., Helg, C., Loliger, C. C., Siren, M. K., Roosnek, E. & Jeannet, M. (1996) Blood 87, 4455–4462. [PubMed] [Google Scholar]
- 8.Santamaria, P., Reinsmoen, N. L., Lindstrom, A. L., Boyce-Jacino, M. T., Barbosa, J. J., Faras, A. J., McGlave, P. B. & Rich, S. S. (1994) Blood 83, 280–287. [PubMed] [Google Scholar]
- 9.Petersdorf, E. W., Kollman, C., Hurley, C. K., Dupont, B., Nademanee, A., Begovich, A. B., Weisdorf, D. & McGlave, P. (2001) Blood 98, 2922–2929. [DOI] [PubMed] [Google Scholar]
- 10.Petersdorf, E. W., Longton, G. M., Anasetti, C., Mickelson, E. M., McKinney, S. K., Smith, A. G., Martin, P. J. & Hansen, J. A. (1997) Blood 89, 1818–1823. [PubMed] [Google Scholar]
- 11.Wallny, H. J. & Rammensee, H. G. (1990) Nature 343, 275–278. [DOI] [PubMed] [Google Scholar]
- 12.Kwiatkowski, D. (2000) Intensive Care Med. 26, Suppl. 1, S89–S97. [DOI] [PubMed] [Google Scholar]
- 13.Rocha, V., Franco, R. F., Porcher, R., Bittencourt, H., Silva, W. A., Jr., Latouche, A., Devergie, A., Esperou, H., Ribaud, P., Socie, G., et al. (2002) Blood 100, 3908–3918. [DOI] [PubMed] [Google Scholar]
- 14.Hill, G. R., Teshima, T., Gerbitz, A., Pan, L., Cooke, K. R., Brinson, Y. S., Crawford, J. M. & Ferrara, J. L. (1999) J. Clin. Invest. 104, 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shizuru, J. A., Jerabek, L., Edwards, C. T. & Weissman, I. L. (1996) Biol. Blood Marrow Transplant. 2, 3–14. [PubMed] [Google Scholar]
- 16.Gandy, K. L., Domen, J., Aguila, H. & Weissman, I. L. (1999) Immunity 11, 579–590. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman, C. L., Colson, Y. L., Wren, S. M., Watkins, S., Simmons, R. L. & Ildstad, S. T. (1994) Blood 84, 2436–2446. [PubMed] [Google Scholar]
- 18.Martin, P. J. (1993) J. Exp. Med. 178, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morse, H. C. (1978) in Origins of Inbred Mice, ed. Morse, H. C. (Academic, New York), pp. 3–21.
- 20.Shimkin, M. B. (1975) Cancer Res. 35, 1597–1598. [PubMed] [Google Scholar]
- 21.Strong, L. C. (1978) in Origins of Inbred Mice, ed. Morse, H. C. (Academic, New York), pp. 45–67.
- 22.Furth, J. (1978) in Origins of Inbred Mice, ed. Morse, H. C. (Academic, New York), pp. 69–97.
- 23.Spangrude, G. J., Heimfeld, S. & Weissman, I. L. (1988) Science 241, 58–62. [DOI] [PubMed] [Google Scholar]
- 24.Cetkovic-Cvrlje, M., Roers, B. A., Waurzyniak, B., Liu, X. P. & Uckun, F. M. (2001) Blood 98, 1607–1613. [DOI] [PubMed] [Google Scholar]
- 25.Dietrich, W., Katz, H., Lincoln, S. E., Shin, H. S., Friedman, J., Dracopoli, N. C. & Lander, E. S. (1992) Genetics 131, 423–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurley, C. K., Wade, J. A., Oudshoorn, M., Middleton, D., Kukuruga, D., Navarrete, C., Christiansen, F., Hegland, J., Ren, E. C., Andersen, I., et al. (1999) Tissue Antigens 53, 394–406. [DOI] [PubMed] [Google Scholar]
- 27.Uchida, N., Tsukamoto, A., He, D., Friera, A. M., Scollay, R. & Weissman, I. L. (1998) J. Clin. Invest. 101, 961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen, J. L. & Weissman, I. L. (2001) Proc. Natl. Acad. Sci. USA 98, 14541–14546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shizuru, J. A., Weissman, I. L., Kernoff, R., Masek, M. & Scheffold, Y. C. (2000) Proc. Natl. Acad. Sci. USA 97, 9555–9560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korngold, R. & Sprent, J. (1983) Immunol. Rev. 71, 5–29. [DOI] [PubMed] [Google Scholar]
- 31.Klein, J. (1986) in Natural History of the Histocompatibility Complex (Wiley, New York), pp. 233–289.
- 32.Marsh, S. G., Bodmer, J. G., Albert, E. D., Bodmer, W. F., Bontrop, R. E., Dupont, B., Erlich, H. A., Hansen, J. A., Mach, B., Mayr, W. R., et al. (2001) Tissue Antigens 57, 236–283. [DOI] [PubMed] [Google Scholar]
- 33.Pullen, J. K., Horton, R. M., Cai, Z. L. & Pease, L. R. (1992) J. Immunol. 148, 953–967. [PubMed] [Google Scholar]
- 34.Mathis, D. J., Benoist, C., Williams, V. E., 2nd, Kanter, M. & McDevitt, H. O. (1983) Proc. Natl. Acad. Sci. USA 80, 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larhammar, D., Hammerling, U., Denaro, M., Lund, T., Flavell, R. A., Rask, L. & Peterson, P. A. (1983) Cell 34, 179–188. [DOI] [PubMed] [Google Scholar]
- 36.Malarkannan, S., Shih, P. P., Eden, P. A., Horng, T., Zuberi, A. R., Christianson, G., Roopenian, D. & Shastri, N. (1998) J. Immunol. 161, 3501–3509. [PubMed] [Google Scholar]
- 37.Choi, E. Y., Christianson, G. J., Yoshimura, Y., Sproule, T. J., Jung, N., Joyce, S. & Roopenian, D. C. (2002) Immunity 17, 593–603. [DOI] [PubMed] [Google Scholar]
- 38.Klein, J. (1986) in Natural History of the Histocompatibility Complex (Wiley, New York), pp. 609–635.
- 39.Melvold, R. W., Wang, K. & Kohn, H. I. (1997) Immunogenetics 47, 44–54. [DOI] [PubMed] [Google Scholar]
- 40.Morse, H. C. (1981) in The Mouse in Biomedical Research, eds. Foster, H. L., Small, J. D. & Fox, J. G. (Academic, New York), pp. 3–16.