Abstract
Epstein–Barr virus (EBV) persists lifelong in infected hosts despite the presence of antiviral immunity. Many viral antigens are expressed during lytic infection. Thus, for EBV to spread, it must have evolved effective ways to evade immune recognition. Here, we report that HLA class II-restricted antigen presentation to T helper cells is hampered in the presence of the lytic-phase protein gp42. This interference with T cell activation involves association of gp42 with class II peptide complexes. Using HLA-DR tetramers, we identify a block in T cell receptor (TCR)–class II interactions imposed by gp42 as the underlying mechanism. EBV gp42 sterically clashes with TCR Vα-domains as visualized by superimposing the crystal structures for gp42–HLA-DR1 and TCR–MHC class II complexes. Blocking TCR recognition provides a previously undescribed strategy for viral immune evasion.
Herpesviruses can persist for life in an immunocompetent host. To withstand antiviral immunity, herpesviruses are equipped with ingenious immunoevasins (1). Most immuneevasion strategies described to date allow viruses to escape from MHC class I-restricted cytotoxic T cells (CTL). Every step of the class I processing and presentation pathway appears to be thwarted by one or more herpesviruses (reviewed in ref. 1). In contrast, much less is known about viral interference with MHC class II-restricted immune responses.
Expression of MHC class II molecules is generally limited to cells with a specialized role in antigen presentation, although other cell types can be induced to express them. At the cell surface, mature MHC class II αβ-heterodimers present peptides to T cell receptors (TCRs) on CD4+ T helper cells (2). T helper cells play a central role in antiviral immunity: they are essential for the induction and maintenance of CTL responses, can exert cytotoxic activity, and provide help for humoral immune responses. In view of the importance of MHC class II-restricted T cells, viruses might also corrupt this part of the immune system.
Escape from T helper cell immunity would be particularly advantageous for Epstein–Barr virus (EBV), which persists in B lymphocytes (3) that constitutively express MHC class II molecules. EBV is a ubiquitous γ-herpesvirus carried by >90% of the adult human population. Primary infection is frequently asymptomatic but can result in infectious mononucleosis. In either case, EBV persists in a latent form under control of the immune response and occasionally reactivates to produce infectious virus. In immunocompromised individuals, the virus can cause life-threatening lymphoproliferative disease, which emphasizes the major contribution of the immune system in controlling viral infection. EBV is also associated with several malignancies, among which are Burkitt's lymphoma and nasopharyngeal carcinoma (reviewed in ref. 3).
Expression of EBV proteins in latently infected cells is limited. All but one of those expressed are targets for CTL responses (4), with the notable exception of Epstein–Barr nuclear antigen 1 (EBNA1), the only protein required for persistence of the viral episome. EBNA1 escapes from MHC class I-restricted presentation to CTL by blocking its proteasomal degradation and, thereby, the generation of antigenic peptides (reviewed in refs. 5 and 6). In contrast, during lytic infection, ≈100 EBV proteins are synthesized for the production of infectious virus, which creates a wide variety of targets for detection and destruction by the immune system. Indeed, T cell immunity against these lytic antigens is present in peripheral blood (7, 8). Evolution of specific evasive strategies to circumvent such existing host immunity might then be anticipated to allow assembly and spread of viral progeny.
EBV is transmitted by saliva, and initial infection of B lymphocytes is thought to occur in the mucosa of the oropharynx. Virions attach to B cells through binding of the main envelope protein gp350 to cellular CD21. Gp42 functions as a cofactor for B cell infection (9). Human MHC (HLA) class II molecules on the host cell serve as interaction partners for gp42 (10, 11).
In this study, we investigated the immunological consequences of EBV gp42–HLA class II interactions. Spriggs et al.(10) suggested that gp42 might interfere with antigen presentation, because they observed a reduction in primary proliferation and cytotoxicity of peripheral blood mononuclear cells in response to recall antigen on addition of a recombinant truncated fusion protein, gp42.Fc. However, these first data were not solidified, cellular expression of full-length gp42 was not achieved, and no mechanism for immune evasion was elucidated. Here, we report that endogenously expressed EBV gp42 can function as a class II immune-evasion molecule. Gp42 binds to HLA class II–peptide complexes, thereby creating a new complex that is impaired for antigen recognition by T helper cells. Gp42 thus offers a window for undetected virus production.
Methods
Cell Lines and Retroviral Transduction. Cell lines were maintained in RPMI medium 1640 with 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 8% FCS (Greiner, Nurtingen, Germany). Mel JuSo (MJS) is a human melanoma cell that expresses HLA-DR3 at the cell surface (12). Transductions with replication-deficient recombinant retroviruses were performed by using the LZRS vector and the φNX-A producer cells as described (13). A retroviral construct was generated that encodes EBV gp42 and a marker, enhanced GFP, separated by an internal ribosome entry site (IRES) to achieve coexpression of both genes. Parental MJS cells were transduced with control IRES-GFP or with gp42-IRES-GFP retroviruses. Continuous and homogenous expression of GFP in the transduced cells was ensured by selection of the GFP+ cells by a FACSVantage cell sorter (Becton Dickinson).
Abs. Abs used to detect EBV gp42 were mAb F-2-1 (14) and rabbit serum no. 32 against amino acids 61–77 of gp42 (M.E.R., D.v.L., and E.J.H.J.W., unpublished data). The following mAbs were used: Tü36 and L243 against HLA-DR αβ complexes; DA6.147 and HB10A against isolated HLA-DR α and β chains, respectively; 16.23 against an HLA-DM-induced conformation of DR3 (15); anti-invariant chain (Ii), VicY1 specific for the cytosolic N terminus, Bü45 for the C terminus; CerCLIP against class II-associated Ii peptide (CLIP)-containing HLA-DR; controls, W6/32 specific for HLA class I molecules and 66IG10 recognizing human transferrin receptor (TfR).
Flow Cytometry. Cell surface expression of specific molecules was determined by indirect immunofluorescence by using the primary Abs listed above and, as a second step, goat-anti-mouse Ig-phycoerythrin (Jackson ImmunoResearch). Cells were analyzed on a FACSCalibur (Becton Dickinson) by using cellquest software.
Biochemical Analysis: Immunoprecipitation, SDS/PAGE, and Western Blotting. In iodination experiments, 10·106 MJS cells were surface-labeled with 1 mCi (1 Ci = 37 GBq) of Na[125I] in PBS by lactoperoxidase-catalyzed iodination. Pulse–chase experiments were essentially performed as described (16). Cells (10–20·106) were metabolically labeled with [35S]methionine (250 μCi/ml; 35S Redivue Promix, Amersham Biosciences) (pulse) followed by a chase in the absence of label. Immunoprecipitations from precleared Nonidet P-40 cell lysates were prepared for SDS/PAGE by either incubation for at least 1 h at 37°C in nonreducing sample buffer (2% SDS/30 mM Tris, pH 6.8/5% glycerol/0.05% bromophenol blue) or boiling for 5 min in reducing sample buffer (2% SDS/50 mM Tris, pH 8.0/10% glycerol/5% 2-mercaptoethanol/0.05% bromophenol blue). Gels were exposed to a phosphor-imaging screen and analyzed with quantity one software (Bio-Rad). Western blot analysis was performed on boiled immunoprecipitates to identify the nature of coprecipitating proteins; total cell lysates were analyzed in parallel. Western blots were stained with HLA-DRα or β-chain-specific mAbs followed by horseradish peroxidase-conjugated goat-anti-mouse Igs (Jackson ImmunoResearch) and visualized by ECLplus (Amersham Biosciences).
Functional T Cell Assays. The HLA-DR3-restricted T helper cell clones R30.95 and M2.11 are specific for a human adenovirus type 5 (Ad5) structural protein (17). T cell recognition was compared for antigen-presenting cells (APC) that were infected with Ad5 for 20 h at a multiplicity of infection of 10 or were mock infected. T cell clone Rp15.1.1 recognizes the Mycobacterium tuberculosis hsp65-derived epitope p3-15 in the context of HLA-DR3 (18). Synthetic p3-15 peptide (2.5 μM) or recombinant hsp65 protein (10 μg/ml) was used as stimulatory antigen for clone Rp15.1.1.
In 96-well round-bottom microtiter plates, 20,000 Ad5-specific or 10,000 M. tuberculosis-specific responder T cells were incubated with 5,000 or 1,000, respectively, irradiated (8,000 rad) MJS, MJS/gfp, or MJS/gp42 cells in triplicate. After 3 days of incubation, supernatants were collected to determine IFN-γ-secretion by using ELISA, and [3H]thymidine was added for an additional 16 h to measure proliferation of T cells. Statistical significance of differences between proliferative responses on stimulation with specific antigens for control and gp42-expressing cells was analyzed by the independent samples Student's t test (two-tailed).
Recombinant gp42 and HLA Tetramers. Recombinant gp42 (rgp42) contains the extracellular domain of the EBV protein (amino acids 33–223) expressed in insect cells (19). Allophycocyanin-labeled MHC class II tetramers were generated as follows. An HLA-DRB1*0401 β-chain construct [composed of the signal peptide of the β-chain, a BamHI-cloning site, a (GGS)3 linker, and amino acids GDTRPR... ARSESA of the extracellular domain of the β-chain, followed by a (GGS)2 linker, basic leucine zipper, and a biotag] was assembled as described (20). The BamHI site was used to insert the influenza virus hemagglutinin (HA)307–319 peptide-encoding sequence. An HLA-DRA*0101 α-chain construct [composed of the signal peptide and extracellular domain of the α-chain (truncated after N217), an acid leucine zipper, and a histidine tag] was a kind gift of D. van Baarle and C. Bronke (Sanquin Research, Amsterdam). HLA class II αβ-heterodimers were produced in insect cells and converted to tetramers as described (20). HLA-A2 tetramers containing the influenza virus matrix (M)58–66 peptide (21) were used as controls.
Tetramer Staining of Human Polyclonal T Cells. Peripheral blood mononuclear cells from an HLA-A2+ and DRB*0401+ (DR4) donor were used to generate influenza virus-specific T cell lines according to Novak et al. (22) with minor modifications. APC consisted of adherent cells that were loaded with 10 μg/ml synthetic peptide representing either the influenza virus DR4-restricted HA307–319 epitope or the A2-restricted M58–66 epitope. As a source of responder T cells, nonadherent cells were labeled with 5 μM 5-(and-6)-carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes) for 10 min at 37°C and were added to the peptide-pulsed APC at a density of 2.5·106 cells per well. After 7 days, nonadherent cells were restimulated with peptide-loaded APC and supplemented with 20 units/ml IL-2. At day 15, responding T cells were harvested and stained with 2 μg/ml DR4/HA307–319 or A2/M58–66 tetramers that were or were not preincubated with the indicated concentrations of soluble rgp42 for 2.5 h on ice. Tetramer staining was performed in combination with phycoerythrin-labeled anti-CD4 or anti-CD8 Abs (Becton Dickinson) and analyzed by flow cytometry. Living cells were gated on the basis of propidium iodide exclusion, and proliferating cells were selected by loss of CFSE staining.
Results
EBV gp42 Associates with HLA-DR Molecules When Expressed by MJS Cells. The aim of the current study was to investigate whether and how EBV gp42 interferes with HLA class II-restricted T cell immunity. With this purpose, we generated an HLA class II+ cell line that endogenously expresses gp42. MJS cells were transduced with a retrovirus encoding gp42. Simultaneous expression of gp42 and GFP was achieved by use of an IRES sequence. A homogeneous GFP+ cell population was selected by flow cytometry and was found to express EBV gp42 at the cell surface (MJS/gp42 cells; data not shown).
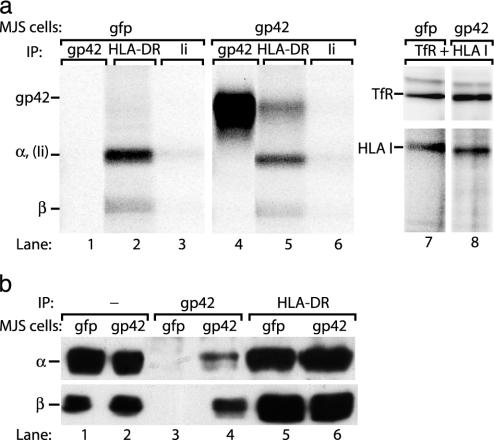
The gp42 gene, BZLF2, encodes a type II transmembrane glycoprotein of 223 amino acids. Surface iodination followed by immunoprecipitation of gp42 with mAb F-2-1 revealed the mature full-length viral protein in MJS/gp42 cells, whereas it was absent from control cells (Fig. 1a, compare lanes 1 and 4). Next, we analyzed whether expression of EBV gp42 in HLA class II+ cells leads to association of gp42 and class II molecules. Immunoprecipitation of HLA-DR αβ complexes showed that class II α and β chains were present at the membrane of iodinated gp42– control cells (Fig. 1a, lane 2). In MJS/gp42 cells, however, an additional protein migrating at the same rate as gp42 coprecipitated with class II complexes (Fig. 1a, compare lanes 4 and 5). No proteins coprecipitated with HLA class I molecules (Fig. 1a, lane 8). These data indicate that gp42 occurs at the surface of MJS/gp42 cells both as free gp42 and in gp42–HLA class II complexes. To confirm the identity of the coprecipitating protein, we performed Western blot analysis of the immune complexes. mAb F-2-1 cannot detect gp42–HLA class II complexes because it recognizes (and blocks) the region of gp42 involved in class II binding (9). Therefore, we generated a rabbit serum (no. 32) against amino acids 61–77 of gp42 to precipitate class II-complexed gp42. On Western blots, total cell lysates were compared with immunoprecipitates obtained with serum no. 32 (against EBV gp42) or mAb Tü36 (against HLA-DRαβ). HLA-DRα and β-chains were detectable in total lysates from MJS/gfp or MJS/gp42 cells (Fig. 1b, lanes 1 and 2). Fig. 2b shows that gp42-immune complexes, retrieved from MJS/gp42 cells, contained both α and β chains (lanes 4). In contrast, no proteins were precipitated from MJS/gfp cells by serum no. 32 (lanes 3), whereas comparable amounts of class II molecules were present in Tü36 precipitates from either cell line (Fig. 1b, lanes 5 and 6). From these experiments, we conclude that EBV gp42 is expressed in MJS/gp42 cells and associates with HLA class II αβ-peptide complexes.
Fig. 1.
Endogenously expressed EBV gp42 associates with HLA class II molecules. (a) Surface-exposed proteins on MJS/gp42 (lanes 4–6 and 8) and control MJS/gfp (lanes 1–3 and 7) cells were labeled with 125I for 1 h. Specific proteins were isolated from cell lysates by immunoprecipitation (IP) with mAbs against gp42 (F-2-1), HLA-DR (Tü36), HLA class I (W6/32), and TfR (66IG10); Ii (VicY1) served as a negative control. Precipitated proteins were separated under nonreducing conditions by SDS/12% PAGE. (b) Total lysates of MJS/gp42 (lanes 2, 4, and 6) and control MJS/gfp (lanes 1, 3, and 5) cells were directly loaded (lanes 1 and 2) or were subjected to immunoprecipitations with rabbit serum no. 32 against gp42 (lanes 3 and 4) and mAb Tü36 against HLA-DR complexes (lanes 5 and 6). Total lysates and immune complexes were boiled in reducing (α) or nonreducing (β) sample buffer, separated by SDS/12% PAGE, and blotted onto poly(vinylidene difluoride) membranes. Western blots were stained with mAbs specific for HLA-DRα (DA6.147) and β (HB10A) chains and visualized by enhanced chemiluminescence.
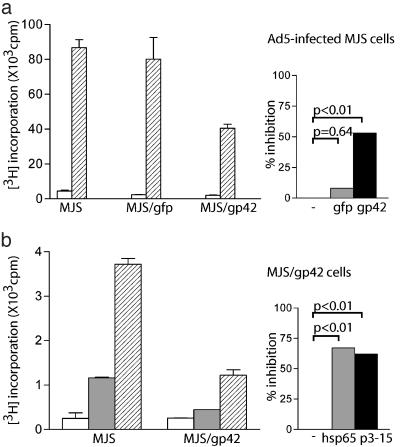
Fig. 2.
Inhibition of T helper responses on antigen presentation by EBV gp42-expressing cells. MJS/gp42 and control cells were assayed for HLA-DR3 presentation of Ad5 ( ) to R30.95 T cells (a) or the M. tuberculosis hsp65 protein (
) to R30.95 T cells (a) or the M. tuberculosis hsp65 protein ( ) and the related p3-15 peptide (
) and the related p3-15 peptide ( ) to Rp15.1.1 T cells (b). [3H]Thymidine incorporation is depicted with error bars for triplicates. (Right) Percentage inhibition of antigen-specific T cell proliferation in response to MJS/gp42 or MJS/gfp cells is depicted compared with (uninfected) MJS cells. Statistical analysis on triplicate wells was performed by the Student's t test, and P values are indicated.
) to Rp15.1.1 T cells (b). [3H]Thymidine incorporation is depicted with error bars for triplicates. (Right) Percentage inhibition of antigen-specific T cell proliferation in response to MJS/gp42 or MJS/gfp cells is depicted compared with (uninfected) MJS cells. Statistical analysis on triplicate wells was performed by the Student's t test, and P values are indicated.
EBV gp42-Expressing Cells Have a Reduced Capacity to Activate T Helper Cells. To assess whether association of EBV gp42 and HLA class II molecules affects activation of T cells, the antigen-presenting capacity of HLA-DR3+ MJS cells was monitored by using well defined HLA-DR3-restricted T cell clones with different specificities. Responsiveness of T cells, R30.95 (Fig. 2a), and M2.11 (data not shown), directed against a structural protein of Ad5 (17), was analyzed after stimulation with Ad5-infected MJS, MJS/gfp, and MJS/gp42 cells. Whereas transduction of MJS cells with control GFP retrovirus did not alter T cell activation (P = 0.64), proliferation induced by virally infected cells expressing gp42 was significantly diminished (P < 0.01, 53% inhibition) (Fig. 2a).
T cell clone Rp15.1.1 recognizes an epitope corresponding to residues 3–15 from hsp65 of M. tuberculosis (18). The use of this clone allowed us to investigate inhibition of T cell responses for antigens that require endogenous processing for presentation as well as for exogenously added peptide epitopes. A significant reduction in T helper cell proliferation was observed in response both to the recombinant hsp65 protein (P < 0.01, 62% inhibition) and to the synthetic p3-15 peptide (P < 0.01, 67% inhibition) when presented by MJS/gp42 cells (Fig. 2b). Similarly, antigen-specific secretion of IFN-γ by the T helper cells was impaired when the APC expressed gp42 (data not shown).
Taken together, these results demonstrate that EBV gp42 negatively affects HLA class II-restricted antigen presentation, irrespective of whether T helper epitopes are introduced as peptides or are generated from endogenously processed proteins after protein uptake or viral infection.
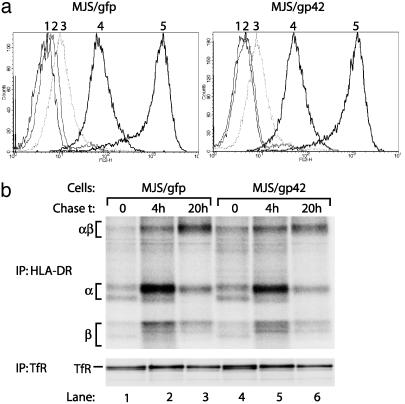
Neither Cell Surface Expression nor Maturation of HLA-DR Is Altered by EBV gp42. To unravel the mechanism by which EBV gp42 impedes T cell recognition, we first determined surface expression levels of HLA-DR molecules by using flow cytometry. Down-regulation of mature HLA–peptide complexes at the cell surface is a T cell evasion strategy frequently observed among herpesviruses (1). However, the presence of EBV gp42 did not alter HLA-DR expression (Fig. 3a). The observed inhibition of T cell activation could also be related to continued association of Ii, or fragments thereof, with the MHC class II αβ-dimers, preventing proper peptide loading. Newly synthesized α and β chains associate with Ii in the endoplasmic reticulum and the αβIi complexes travel from the trans-Golgi network toward acidic compartments where Ii is proteolytically cleaved. The chaperone HLA-DM facilitates replacement of the remaining CLIP fragment by peptides that bind within the class II groove to be displayed at the cell surface (2, 23). As a result of defective intracellular processing, levels of surface-exposed Ii could be increased, but this appeared not to be the case in gp42+ cells (Fig. 3a). Furthermore, both in control and in gp42-expressing MJS cells, HLA-DR3 molecules come into contact with HLA-DM, inducing a conformational change detected by mAb 16.23 (15) and leading to removal of CLIP, as reflected by the absence of surface reactivity with mAb CerCLIP (24) (Fig. 3a).
Fig. 3.
Surface expression and maturation of HLA-DR αβ-peptide complexes is not altered in MJS/gp42 cells. (a) Flow-cytometry histograms are depicted for surface staining of MJS/gfp and MJS/gp42 cells with the following mAbs: CerCLIP for HLA-DR/CLIP complexes (peak 2), Bü45 for Ii (peak 3), 16.23 for an HLA-DM-induced conformation of DR3 (peak 4), and L243 for HLA-DR (peak 5). As a negative control, no primary Ab was added (peak 1). (b) MJS/gp42 (lanes 4–6) and control MJS/gfp (lanes 1–3) cells were metabolically labeled for 1 h with [35S]methionine and chased for 0, 4, and 20 h, followed by cell lysis. HLA-DR complexes were isolated by immunoprecipitation (IP) with mAb L243, were incubated in nonreducing sample buffer with SDS at 37°C, which reveals the SDS-stable class II αβ dimers (αβ), and were analyzed on SDS/12% PAGE. As a control protein, TfR was precipitated by mAb 66IG10 and separated under reducing conditions by SDS/10% PAGE.
We next examined intracellular transport and maturation of HLA class II molecules by following the generation of SDS-stable class II complexes over time. The formation of HLA class IIαβ–peptide complexes that are resistant to dissociation in 2% SDS requires release of Ii and acquisition of peptides by a process that involves HLA-DM (15, 25). The pulse–chase experiment depicted in Fig. 3b indicates that SDS-stable class II complexes were formed to the same extent and with comparable kinetics in MJS/gp42 and control MJS/gfp cells. As a control, consistent amounts of TfR were recovered at all time points studied (Fig. 3b). These combined experiments show that gp42-induced inhibition of antigen-specific T helper cell recognition cannot be explained by a decrease in mature HLA-DRαβ–peptide complexes at the cell surface.
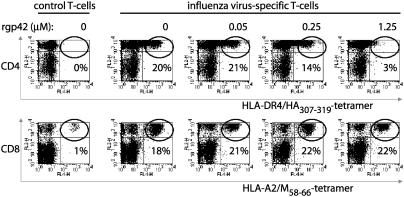
EBV gp42 Blocks TCR–HLA-DR Interactions. As an alternative to interference with antigen processing, viral proteins associated with MHC molecules at the cell surface might inhibit T cell activation by causing a block in TCR engagement. We used MHC/peptide tetramers to assess directly any interference with TCR–HLA interactions by EBV gp42. In these experiments, a soluble recombinant form of gp42 (rgp42) was used that lacks the transmembrane and cytoplasmic domains, but is still capable of binding to HLA class II molecules (19) and impeding T cell activation (data not shown). We tested whether the addition of rgp42 affected staining of influenza virus-specific T cells with HLA-DR4 tetramers containing the viral HA307–319 epitope. Tetramers of HLA-A2 with influenza virus M58–66 were analyzed in parallel as a control for nonspecific effects. Polyclonal influenza virus-specific T cells were generated from a healthy HLA-A2+ and DR4+ donor by stimulation of CFSE-labeled peripheral blood mononuclear cells with M58–66 or HA307–319 peptides. We analyzed the specificity of responding T cells that had divided and had thus become CFSE– (Fig. 4). Few tetramer+ cells were detected when T cells were left unstimulated (control). After peptide stimulation, ≈20% of the responding CD4+ T cells associated through their TCR with the specific DR4/HA307–319 tetramers (Fig. 4 Upper). Comparable amounts of specific CD8+ CFSE– T cells were visualized with A2/M58–66 tetramers (Fig. 4 Lower). The addition of 1.25 μM rgp42 almost abolished TCR-DR4 interactions (staining was reduced to 3%), and this effect was dose-dependent (Fig. 4 Upper). In contrast, HLA class I molecules were normally engaged by TCRs irrespective of the addition of gp42 (Fig. 4 Lower). In conclusion, these data indicate that EBV gp42 reduced T cell recognition of HLA-DR–peptide complexes through a block in TCR engagement.
Fig. 4.
TCR interactions with complexes of peptide–HLA class II, but not class I, are abolished in the presence of soluble EBV gp42. Peripheral blood mononuclear cells from a healthy HLA-A2+ DR4+ donor were labeled with CFSE and stimulated in vitro with DR4/HA307–319 (Upper) or A2/M58–66 (Lower) or without peptides (control). Responding T cells were incubated with specific allophycocyanin-conjugated tetramers in the presence or absence of rgp42. rgp42 concentrations of 0.05, 0.25, and 1.25 μM correspond to gp42:class II ratios of 0.5:1, 3:1, and 12:1, respectively. Cells were stained with phycoerythrin-conjugated mAbs to CD4 (Upper) or CD8 (Lower). Each flow-cytometry dot plot represents ≈45,000 propidium iodide–, CFSE– cells; percent values refer to the percentage of CD4+ or CD8+ T cells that stained with the tetramers (top right quadrant).
Discussion
In this study, we show that EBV gp42 impairs TCR-mediated activation of antigen-specific T helper cells in an HLA class II-dependent manner. Endogenous cellular expression of gp42 results in formation of gp42 HLA-DRαβ–peptide complexes and inhibition of T helper cells specific for various antigens. By using HLA-DR tetramers to directly visualize TCR–class II interactions, we demonstrate that the mechanism of immune evasion mediated by EBV gp42 relies on direct blockage of TCR engagement.
Expression of EBV gp42 in HLA-DR+ cells was achieved by retroviral transduction of the gp42 gene, which ensures stable and homogeneous protein expression in all cells. Our approach facilitates the evaluation of potential immune modulatory effects, but it does not take into account the viral context of gp42. We chose this approach for two reasons. First, no fully permissive culture system for EBV is currently available, which has severely hampered analysis of lytic-cycle gene expression. B lymphocytes transformed in vitro with EBV and EBV+ Burkitt's lymphoma cell lines at large display latent gene expression. In either cell type, a small fraction of cells spontaneously gives rise to virus production, and the efficiency of viral reactivation can be enhanced by cross-linking surface IgG or by treatment with chemical inducers, such as phorbol esters and/or n-butyrate (14, 26, 27). However, EBV proceeds through the full lytic cycle only in a limited number of cells and in a nonsynchronous fashion. Second and more importantly, interference with immune responses in cells supporting viral replication could be the result of proteins other than gp42. EBV encodes several gene products with defined immunomodulatory functions (reviewed in refs. 5 and 6). The viral IL-10 homolog, encoded by BCRF1, suppresses production of the proinflammatory cytokines IFN-γ and IL-12. Macrophage proliferation is inhibited by the BARF1-encoded soluble receptor for CSF-1. A viral bcl-2 homolog (BHRF-1) can confer resistance to apoptosis, e.g., induced in target cells on recognition by CTL. LMP1 enhances antigen-presenting functions in EBV-transformed B-LCL (28, 29), but expression of the immediate early transactivator BZLF1 can completely reverse these effects (30). BZLF1 also inhibits IFN-γ signaling by decreasing its receptor expression (31). Finally, tumor-derived Burkitt's lymphoma cell lines frequently have impaired antigen-processing and presenting capacities (28). For the identification of most herpesvirus-encoded immunoevasins and definition of the molecular basis of their function, cell lines that stably express the viral protein of interest have been instrumental.
EBV gp42 was initially identified as a cofactor for B cell infection. On the viral envelope, gp42 occurs in a complex with two other viral glycoproteins, gH and gL (9, 14). Association with gH–gL is not required for folding and transport of gp42 (14). Here, we demonstrated that in the absence of its viral partners, gp42 is expressed at the cell surface of MJS/gp42 cells (Fig. 1). HLA class II molecules serve as receptors for gp42 on B lymphocytes (9–11). The indispensability of gp42–class II interactions for viral entry has been demonstrated by several findings. A mutant virus lacking gp42 cannot infect or transform B cells (32). Vice versa, HLA class II– cells are not susceptible to EBV infection unless class II expression is restored (9, 11). Moreover, recent experiments in EBV-producing cells indicate that the amount of gp42 incorporated in gH–gL complexes on the virion has a significant effect on viral tropism for cells differing in expression of HLA class II (33).
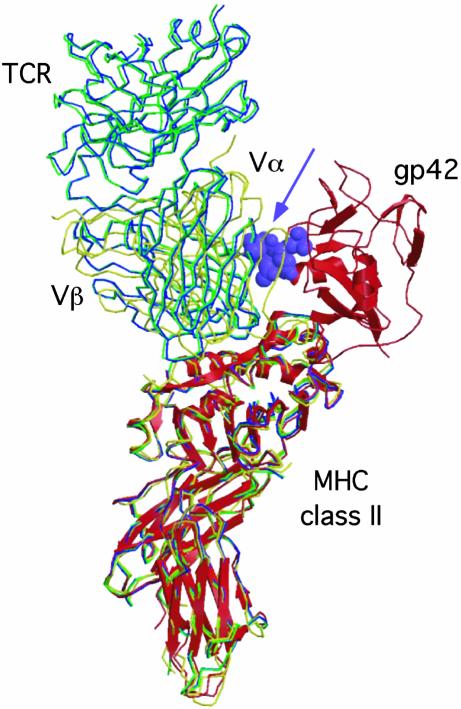
In contrast to the attention given to virological processes, no studies to date have addressed the cellular and immunological consequences of coexpression of EBV gp42 and MHC class II molecules. Here, we report that gp42-expressing cells display a substantial reduction in their capacity to activate well defined class II-restricted T cell clones. EBV gp42 impairs antigen recognition of both peptide epitopes and proteins that require endogenous processing for presentation. Recently, the crystal structure of gp42–HLA-DR1 complexes has been resolved, demonstrating that gp42 is positioned along the side of the HLA–peptide-binding site for the TCR (19). The gp42–HLA-DR structure is reminiscent of superantigen–MHC class II complexes, suggesting that gp42 might cross-link TCR and class II molecules. As opposed to superantigens, however, gp42 cannot associate with TCRs (data not shown). Fig. 5 shows that binding of gp42 may sterically interfere with engagement of MHC class II–peptide complexes for two defined TCRs, HA1.7 (34, 35) and D10 (36), that have been crystallized so far. Some variability, however, appears to occur in the arrangement of different TCR–MHC complexes (37). Furthermore, gp42 binds peripherally to the β-chain rather than completely covering the TCR–HLA interaction site. Hence, the possibility had existed that even gp42-complexed class II molecules could present antigen to specific T cells. Here, the use of HLA-DR tetramers allowed us to show that EBV gp42 blocks TCR–class II interactions. Inhibition of tetramer binding is already observed at a low micromolar concentration of gp42, at which gp42 is present in only a modest stoichiometric excess over MHC class II (Fig. 4), consistent with a high-affinity interaction. Moreover, various TCRs are hindered by gp42, as tetramer binding was blocked for polyclonal T cell responses (Fig. 4). On the basis of our results, we can now conclude that the mechanism of immune evasion relies on interference by gp42 with TCR recognition of MHC class II–peptide complexes. This is consistent with data showing that exogenously added gp42 also inhibits T helper cell activation and explains the inhibitory effects on T cell activation in response to processed antigens and exogenously provided peptide epitopes. Binding of gp42 to the side of the HLA class II–peptide-binding groove implies that peptide content is not likely to influence gp42–class II interactions. This, combined with the observation that gp42 can bind to a wide array of class II alleles (11), makes interference by gp42 with TCR recognition an evasion strategy that would allow undetected viral propagation in a large population of class II-diverse human hosts.
Fig. 5.
EBV gp42 sterically clashes with the TCR in known TCR–MHC class II structures. Superposition of the gp42–DR1 structure (shown as red ribbons) with the crystal structures of the HA1.7-TCR–DR1 complex (green), the HA1.7-TCR–DR4 complex (blue), and the D10-TCR–I-Ak complex (yellow). The gp42 loop including residues 157–161 clashes with the TCR Vα domains and is shown as purple Corey–Pauling–Koltun model atoms (arrow). The D10-TCR has the greatest overlap with gp42, whereas the HA1.7 structures are rotated slightly away from gp42.
Evasion from CD4+ T cell immunity is particularly relevant for viruses, such as EBV, that infect MHC class II+ host cells. EBV-specific CD4+ T helper responses have not been examined in the same detail as the CD8+ T cell response (4), but recent data indicate that ubiquitous CD4+ T cell responses specific for EBV are present and display direct antiviral capacities (38–40). The critical role for CD4+ T cells in controlling chronic infection has been demonstrated in mice infected with the murine γ-herpesvirus MHV-68 (41). A similar function for human CD4+ T cells in maintaining anti-EBV immunity is suggested by the increased EBV-related morbidity observed in AIDS patients with reduced CD4+ T cell counts (42). It thus appears profitable for EBV to (temporarily) impair MHC class II-restricted CD4+ T cell recognition.
Few data are available on interference with HLA class II-restricted antigen presentation after viral infection. Two human cytomegalovirus gene products are reported to affect MHC class II expression. US2 targets the class II DR-α and DM-α molecules for degradation by proteasomes shortly after synthesis (43), and US3 prevents peptide loading of class II complexes through inefficient sorting to the MHC class II-loading compartment (44). The HIV Nef protein causes an increase in surface expression of immature class II αβIi complexes and a reduction in peptide presentation to T cells (45).
In this study, we have shown that EBV may use yet another strategy: gp42 inhibits antigen-specific activation of T helper cells by binding to HLA class II molecules and blocking TCR engagement.
Acknowledgments
We thank Drs. J. W. Drijfhout, G. Nolan, M. Heemskerk, A. Ziegler, P. Cresswell, and W. Knapp for providing materials. Drs. R. Toes and K. Melief are acknowledged for critically reading the manuscript. This study was financially supported by the Dutch Cancer Society (Grant RUL 98-179) and the Netherlands Organization for Scientific Research.
Abbreviations: Ad5, human adenovirus type 5; APC, antigen-presenting cells; CFSE, 5-(and-6)-carboxyfluorescein diacetate succinimidyl ester; CLIP, class II-associated invariant chain peptide; CTL, cytotoxic T cells; EBV, Epstein–Barr virus; Ii, invariant chain; HA, hemagglutinin; IRES, internal ribosome entry site; MJS, Mel JuSo; TCR, T cell receptor; TfR, transferrin receptor.
References
- 1.Tortorella, D., Gewurz, B. E., Furman, M. H., Schust, D. J. & Ploegh, H. L. (2000) Annu. Rev. Immunol. 18, 861–926. [DOI] [PubMed] [Google Scholar]
- 2.Cresswell, P. (1994) Annu. Rev. Immunol. 12, 259–293. [DOI] [PubMed] [Google Scholar]
- 3.Rickinson, A. B. & Kieff, E. (2001) in Field's Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadelphia), pp. 2575–2627.
- 4.Rickinson, A. B. & Moss, D. J. (1997) Annu. Rev. Immunol. 15, 405–431. [DOI] [PubMed] [Google Scholar]
- 5.Levitsky, V. & Masucci, M. G. (2002) Virus Res. 88, 71–86. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, J. I. (1999) Curr. Opin. Immunol. 11, 365–370. [DOI] [PubMed] [Google Scholar]
- 7.Hislop, A. D., Annels, N. E., Gudgeon, N. H., Leese, A. M. & Rickinson, A. B. (2002) J. Exp. Med. 195, 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steven, N. M., Annels, N. E., Kumar, A., Leese, A. M., Kurilla, M. G. & Rickinson, A. B. (1997) J. Exp. Med. 185, 1605–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, Q., Spriggs, M. K., Kovats, S., Turk, S. M., Comeau, M. R., Nepom, B. & Hutt-Fletcher, L. M. (1997) J. Virol. 71, 4657–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spriggs, M. K., Armitage, R. J., Comeau, M. R., Strockbine, L., Farrah, T., Macduff, B., Ulrich, D., Alderson, M. R., Mullberg, J. & Cohen, J. I. (1996) J. Virol. 70, 5557–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haan, K. M. & Longnecker, R. (2000) Proc. Natl. Acad. Sci. USA 97, 9252–9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Ham, S. M., Tjin, E. P., Lillemeier, B. F., Gruneberg, U., van Meijgaarden, K. E., Pastoors, L., Verwoerd, D., Tulp, A., Canas, B., Rahman, D., et al. (1997) Curr. Biol. 7, 950–957. [DOI] [PubMed] [Google Scholar]
- 13.Heemskerk, M. H., Hooijberg, E., Ruizendaal, J. J., van der Weide, M. M., Kueter, E., Bakker, A. Q., Schumacher, T. N. & Spits, H. (1999) Cell Immunol. 195, 10–17. [DOI] [PubMed] [Google Scholar]
- 14.Li, Q., Turk, S. M. & Hutt-Fletcher, L. M. (1995) J. Virol. 69, 3987–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verreck, F. A., Fargeas, C. A. & Hammerling, G. J. (2001) Eur. J. Immunol. 31, 1029–1036. [DOI] [PubMed] [Google Scholar]
- 16.Wiertz, E. J., Jones, T. R., Sun, L., Bogyo, M., Geuze, H. J. & Ploegh, H. L. (1996) Cell 84, 769–779. [DOI] [PubMed] [Google Scholar]
- 17.Heemskerk, B., Veltrop-Duits, L. A., Van Vreeswijk, T., Ten Dam, M. M., Heidt, S., Toes, R. E., Van Tol, M. J. & Schilham, M. W. (2003) J. Virol. 77, 6562–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geluk, A., van Meijgaarden, K. E., Janson, A. A., Drijfhout, J. W., Meloen, R. H., de Vries, R. R. & Ottenhoff, T. H. (1992) J. Immunol. 149, 2864–2871. [PubMed] [Google Scholar]
- 19.Mullen, M. M., Haan, K. M., Longnecker, R. & Jardetzky, T. S. (2002) Mol. Cell 9, 375–385. [DOI] [PubMed] [Google Scholar]
- 20.Schepers, K., Toebes, M., Sotthewes, G., Vyth-Dreese, F. A., Dellemijn, T. A., Melief, C. J., Ossendorp, F. & Schumacher, T. N. (2002) J. Immunol. 169, 3191–3199. [DOI] [PubMed] [Google Scholar]
- 21.Altman, J. D., Moss, P. A., Goulder, P. J., Barouch, D. H., McHeyzer-Williams, M. G., Bell, J. I., McMichael, A. J. & Davis, M. M. (1996) Science 274, 94–96. [DOI] [PubMed] [Google Scholar]
- 22.Novak, E. J., Liu, A. W., Nepom, G. T. & Kwok, W. W. (1999) J. Clin. Invest. 104, R63–R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pieters, J. (1997) Curr. Opin. Immunol. 9, 89–96. [DOI] [PubMed] [Google Scholar]
- 24.Denzin, L. K., Robbins, N. F., Carboy-Newcomb, C. & Cresswell, P. (1994) Immunity 1, 595–606. [DOI] [PubMed] [Google Scholar]
- 25.Neefjes, J. J. & Ploegh, H. L. (1992) EMBO J. 11, 411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe, M., Lear, A. L., Croom-Carter, D., Davies, A. H. & Rickinson, A. B. (1992) J. Virol. 66, 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takada, K. & Ono, Y. (1989) J. Virol. 63, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowe, M., Khanna, R., Jacob, C. A., Argaet, V., Kelly, A., Powis, S., Belich, M., Croom-Carter, D., Lee, S., Burrows, S. R., et al. (1995) Eur. J. Immunol. 25, 1374–1384. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Q., Brooks, L., Busson, P., Wang, F., Charron, D., Kieff, E., Rickinson, A. B. & Tursz, T. (1994) Eur. J. Immunol. 24, 1467–1470. [DOI] [PubMed] [Google Scholar]
- 30.Keating, S., Prince, S., Jones, M. & Rowe, M. (2002) J. Virol. 76, 8179–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison, T. E., Mauser, A., Wong, A., Ting, J. P. & Kenney, S. C. (2001) Immunity 15, 787–799. [DOI] [PubMed] [Google Scholar]
- 32.Wang, X. & Hutt-Fletcher, L. M. (1998) J. Virol. 72, 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borza, C. M. & Hutt-Fletcher, L. M. (2002) Nat. Med. 8, 594–599. [DOI] [PubMed] [Google Scholar]
- 34.Hennecke, J., Carfi, A. & Wiley, D. C. (2000) EMBO J. 19, 5611–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hennecke, J. & Wiley, D. C. (2002) J. Exp. Med. 195, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinherz, E. L., Tan, K., Tang, L., Kern, P., Liu, J., Xiong, Y., Hussey, R. E., Smolyar, A., Hare, B., Zhang, R., et al. (1999) Science 286, 1913–1921. [DOI] [PubMed] [Google Scholar]
- 37.Hennecke, J. & Wiley, D. C. (2001) Cell 104, 1–4. [DOI] [PubMed] [Google Scholar]
- 38.Khanna, R., Burrows, S. R., Thomson, S. A., Moss, D. J., Cresswell, P., Poulsen, L. M. & Cooper, L. (1997) J. Immunol. 158, 3619–3625. [PubMed] [Google Scholar]
- 39.Leen, A., Meij, P., Redchenko, I., Middeldorp, J., Bloemena, E., Rickinson, A. & Blake, N. (2001) J. Virol. 75, 8649–8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munz, C., Bickham, K. L., Subklewe, M., Tsang, M. L., Chahroudi, A., Kurilla, M. G., Zhang, D., O'Donnell, M. & Steinman, R. M. (2000) J. Exp. Med. 191, 1649–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardin, R. D., Brooks, J. W., Sarawar, S. R. & Doherty, P. C. (1996) J. Exp. Med. 184, 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Baarle, D., Hovenkamp, E., Callan, M. F., Wolthers, K. C., Kostense, S., Tan, L. C., Niesters, H. G., Osterhaus, A. D., McMichael, A. J., van Oers, M. H., et al. (2001) Blood 98, 146–155. [DOI] [PubMed] [Google Scholar]
- 43.Tomazin, R., Boname, J., Hegde, N. R., Lewinsohn, D. M., Altschuler, Y., Jones, T. R., Cresswell, P., Nelson, J. A., Riddell, S. R. & Johnson, D. C. (1999) Nat. Med. 5, 1039–1043. [DOI] [PubMed] [Google Scholar]
- 44.Hegde, N. R., Tomazin, R. A., Wisner, T. W., Dunn, C., Boname, J. M., Lewinsohn, D. M. & Johnson, D. C. (2002) J. Virol. 76, 10929–10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stumptner-Cuvelette, P., Morchoisne, S., Dugast, M., Le Gall, S., Raposo, G., Schwartz, O. & Benaroch, P. (2001) Proc. Natl. Acad. Sci. USA 98, 12144–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]