Abstract
Hepatocyte nuclear factors 3 α, β, and γ (Foxa-1, -2, and -3) are transcriptional activators of important metabolic genes in the liver that are suppressed by the actions of insulin. Here, we show that the activation of phosphatidylinositol 3-kinase–Akt by insulin induces Foxa-2 phosphorylation, nuclear exclusion, and inhibition of Foxa-2-dependent transcriptional activity. Foxa-2 physically interacts with Akt, a key mediator of the phosphatidylinositol 3-kinase pathway and is phosphorylated at a single conserved site (T156) that is absent in Foxa-1 and Foxa-3 proteins. This Akt phosphorylation site in Foxa-2 is highly conserved from mammals to insects. Mutant Foxa-2T156A is resistant to Akt-mediated phosphorylation, nuclear exclusion, and transcriptional inactivation of Foxa-2-regulated gene expression. These results implicate an evolutionarily conserved mechanism in the regulation of Foxa-2-dependent transcriptional control by extracellular signals such as insulin.
Keywords: phosphoenolpyruvate carboxykinase, Foxo-1, phosphatidylinositol 3-kinase
The hepatocyte nuclear factor 3 (Hnf-3)/forkhead family of transcription factor in mammals include three genes designated as Foxa-1 (Hnf-3α), Foxa-2 (Hnf-3β), and Foxa-3 (Hnf-3γ) (1). These all have in common a highly conserved 100-aa winged-helix motif that is responsible for monomeric recognition of specific DNA target sites (2). Foxa proteins play a central role in maintaining normal glucose homeostasis by regulating gene expression of rate-limiting enzymes of gluconeogenesis and glycogenolysis in the liver and kidney, including phosphoenol-pyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6pc), and by regulating glucagon and Pdx-1 gene expression in pancreatic α and β cells, respectively (3–5).
In the liver, insulin regulates gene expression of enzymes of gluconeogenesis and glycogenolysis by suppressing transcriptional activity. These pathways ensure that hepatic glucose production is suppressed in the fed state (when insulin levels are increased), and glucose levels are maintained in times of starvation (when serum insulin is low and glucagon is increased) (6). The inability of insulin to suppress endogenous glucose production by inadequate insulin secretion in type 1 diabetes or impaired insulin signaling in type 2 diabetes contributes to the pathogenesis of these common metabolic diseases. The mechanisms by which insulin signaling pathways can modulate the activity of key transcription factors involved in the regulation of metabolic genes is incompletely understood.
The serine/threonine kinase PKB/Akt is one downstream target of phosphatidylinositol 3-kinase (PI3-kinase) and plays an important role in mediating effects of insulin on hepatic glucose production, glycogen, and protein synthesis (7–9). Upon activation, Akt is translocated to the nucleus where it exerts effects on gene activity by phosphorylation of target proteins like Gsk3 and Bad (10–12). Genetic studies of the PI3-kinase–Akt signaling pathway in the nematode Caenorhabditis elegans have established that this signaling cascade suppresses the function of the transcription factor daf16, which belongs to the forkhead/ winged-helix family of transcription factors. Mutations in the insulin/Igf-1 receptor homologue (daf-2), the catalytic subunit of PI3-kinase (age-1), or Akt (akt1 and akt2) result in increased longevity and constitutive dauer formation, a stage of developmental arrest and reduced metabolic activity that enhances survival periods of food deprivation and other environmental stresses (13). In each case, mutation of daf-16 restored normal life span and prevented entry into dauer stage (14, 15). Subsequently, studies in mammals have shown that the Fkhr (Foxo-1), Fkhrl1 (Foxo-3), and AFX (Foxo-4) genes, members of the human forkhead family, also constitute downstream targets of Akt (16, 17). For instance, the Foxo-1 protein can be phosphorylated by Akt, which causes repression of transcriptional activity of insulin growth factor binding protein 1 (Igfbp-1), and G6pc (18). Furthermore, genetic studies in mice have provided evidence that downstream components of the insulin/Igf-1 signaling pathway are essential for normal energy homeostasis and growth. Mice lacking Akt2 have an impaired ability of insulin to inhibit glucose production in the liver and muscle (19). In contrast, mice lacking Akt1 have normal glucose homeostasis, but impaired fetal and postnatal growth (20).
In this article, we explore the molecular basis by which Foxa-2-dependent transcription is inhibited by insulin. Our results identify Foxa-2 as a novel target of Akt and indicate that phosphorylation at a single conserved site is both necessary and sufficient to inhibit the transcriptional activity of Foxa-2.
Methods
Materials. Insulin was from Sigma. LY294002 and PD98059 were from Calbiochem.
Generation of Plasmids. Expression vectors for Foxa-1 and Foxa-2 were generated by cloning the coding regions into pcDNA3 either with or without fusion to an N-terminal Flag/hemagglutinin (HA) tag. Mutants (T156A and R153K) were generated by PCR mutagenesis using the QuikChange protocol (Stratagene). Expression vectors for HA-Akt1 (pCMV-HA-Akt) were generated by cloning the coding region of human Akt1 into pcDNA3 fused to an N-terminal HA tag. Expression vector pCMV-HA-Lck-Akt encodes the HA-tagged form of constitutively active Akt and contains the N-terminal localization sequence from the Lck gene, a consensus sequence for both myristoylation and palmitylation (21). Vector pCMV-HA-Akt1K179A encodes an inactive Akt and has been described (21, 22). Bacterial expression vectors of Foxa-2 and Foxa-2T156A were generated by cloning the cDNA into pGEX-4T2 (Amersham Pharmacia).
Cell Lines and Primary Hepatocytes. HepG2 and human embryonic kidney (HEK)293 cells were maintained in DMEM supplemented with 4.5 g/liter glucose, 10% FCS, 2 mM glutamine, and 50 μg/ml gentamycin/streptomycin in a humidified incubator at 5% CO2.
Transfections and Transactivation Assay. HepG2 or HEK293 cells were transfected with reporter genes (p6xCdx-TkLuc or pPEPCK-Luc), pCMV-β-galactosidase, and the expression vectors for WT or mutant Foxa-1 and Foxa-2 and human Akt1/Akt2 by using the transfection reagent Fugene6 (Roche Molecular Biochemicals). Cells were grown for 48 h, and luciferase activity was measured by using the Luciferase Detection System (Promega). Luciferase was normalized for transfection efficiency by β-galactosidase activity.
Expression and Purification of Recombinant GST-Foxa-2. BL21 Escherichia coli cells carrying the expression plasmid for either pGST-Foxa-2 or pGST-Foxa-2T156A were grown to an OD600 of 0.8 and, protein expression was induced by addition of 0.4 mM isopropyl β-d-thiogalactoside. Cells were harvested by centrifugation and lysed in 10 mM Tris·HCl, pH 7.4, and 30 mM NaCl by sonication at 4°C. Proteins were purified by using Mono-Q and Suprose 12 columns with an FPLC system (Amersham Pharmacia). Purity of the protein was determined by SDS/PAGE.
Immunoprecipitation. Foxa-2 and Akt1 were precipitated from cell lysates by using polyclonal anti-Foxa-2 antibodies (23) or monoclonal anti-HA antibody (Sigma) bound to gammabind-Sepharose (Amersham Pharmacia) overnight at 4°C. Proteins were eluted with SDS-loading buffer, separated by 12% SDS/PAGE, and analyzed by Western blotting using either monoclonal anti-Foxa-2 antibody (1:4,000) (Developmental Studies Hybridoma Bank, Iowa City) or polyclonal anti-HA antibody (1:2,000) (Sigma) and secondary antibodies linked to horseradish peroxidase (Calbiochem). Proteins were visualized by using the ECL detection system (NEN).
In Vitro Kinase Assay. Phosphorylation of Foxa-2 was analyzed by using an in vitro kinase assay. Akt was precipitated from 200 μg of total protein lysate from HEK293 cells transfected with pCMV-HA-Akt or pCMV-HA-Lck-Akt by using anti-HA antibody (Sigma) bound to gammabind-Sepharose (Amersham Pharmacia) for 2 h at 4°C. Precipitates were washed three times with kinase buffer [25 mM Mops, pH 7.4/25 mM β-glycerophosphate/20 mM MgCl2/2 mM MnCl2/1 mM DTT/protease inhibitor mixture (Roche Molecular Biochemicals)] and incubated with 5 μg of either purified Foxa-2 and Foxa-2T156A or with GST-crosstide (24) (positive control) in the presence of 0.5 μCi γ-ATP for 15 min at 37°C. Proteins were eluted with SDS-loading buffer, separated by SDS/PAGE (13.5%), and analyzed by autoradiography.
Electrophoretic Mobility-Shift Assay. Whole-cell extracts from transfected HEK293 cells (20 μg) were incubated with a 32P-labeled double-stranded oligonucleotide probe with the Foxa binding sites of the Igfbp-1 promoter (25).
Immunofluorescence Microscopy. Cells were fixed for 30 min at room temperature with 2% paraformaldehyde. For immunofluorescent detection of Foxa-1 or Foxa-2, fixed cells were incubated with respective polyclonal antibodies (1:100) (23) overnight at 4°C. After washing, the cells were treated with anti-rabbit IgG secondary antibody linked to Alexa Fluor 488 (Molecular Probes). Immunofluorescent staining was visualized by using laser-scanning microscopy.
Results
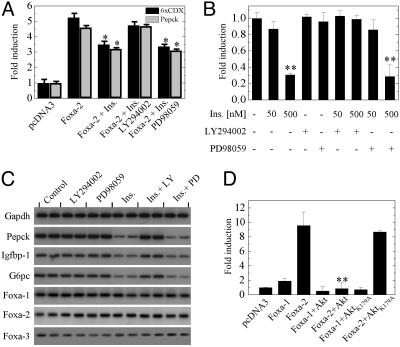
Insulin-Dependant Decrease of Foxa-2 Is Mediated by PI3-Kinase–Akt. To analyze whether Foxa-2 activity is regulated by insulin, we transfected HepG2 cells with an Foxa-2 expression vector and either plasmid pPEPCK-Luc or p6xCdx-TkLuc that contains a 621-bp promoter fragment of the human PEPCK gene or six Foxa binding sites of the murine Cdx-2 gene upstream of a minimal promoter and the luciferase gene, respectively (26). Cotransfection of Foxa-2 with both reporter constructs led to a ≈5-fold increase in activity compared with control transfection. Treatment of the cells with insulin for the duration of transfection significantly decreased Foxa-2 activity. Because insulin stimulation has been shown to activate mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK) and PI3-kinase signaling we tested the effect of inhibitors of both pathways. Coincubation with the PI3-kinase inhibitor LY294002 (10 μM) prevented insulin-mediated decrease of Foxa-2 activity, whereas the MEK inhibitor PD98059 (10 μM) did not influence insulin regulation of Foxa-2 activity (Fig. 1A).
Fig. 1.
Insulin decreases Foxa-2 activity via PI3-kinase–Akt activation. (A) HepG2 cells were transfected with an expression vector for Foxa-2 and pPEPCK-Luc (gray bars) or p6xCdx-TkLuc (black bars). Cells were treated with 100 nM insulin, 10 μM LY294002, or 10 μM PD98059 alone or in combination. (B) HepG2 cells were transfected with p6xCdx-TkLuc. Cells were treated with insulin, 10 μM LY294002, or 10 μM PD98059 at the indicated concentrations, alone or in combination. (C) RT-PCR analysis of Foxa-1–3 and target genes. Primary hepatocytes were grown in the absence and presence of 50 nM insulin, LY294002, and PD98059 for 6 h before gene expression analysis. (D) HepG2 cells were transfected with an expression vector for Foxa-1, Foxa-2, Akt, or AktK179A alone or in combination. p6xCdx-TkLuc was used as reporter gene. In all experiments luciferase activity was normalized to β-galactosidase activity. All vectors were transfected at 125 ng. Values are mean of six independent experiments ± SD. *, P ≤ 0.01; **, P ≤ 0.001.
We next investigated whether insulin decreases endogenous Foxa-2 activity in HepG2 cells. Cells were transfected with the reporter construct alone, and Foxa-2 activity was measured after stimulation with insulin and PI3-kinase or MEK inhibitors. Insulin stimulation led to a dose-dependent decrease of Foxa-2 activity (80% decrease at 500 nM insulin). This decrease in activity was ablated when cells were coincubated with LY294002 but not with PD98059 (Fig. 1B).
To analyze whether inhibition of Foxa-2 target gene expression by insulin is controlled at a transcriptional or posttranscriptional level, we measured mRNA levels of Pepck, G6pc, and Igfbp-1 in primary hepatocytes that were cultured either in the presence or absence of insulin, PI3-kinase, or MEK inhibitors. We found that mRNA levels were reduced 3- to 4-fold in insulin- or insulin/PD98059-treated hepatocytes but not in controls (no insulin) or insulin/LY294002-treated cells (Fig. 1C). The reduced expression could not be attributed to increased expression levels of Foxa-1, -2, and -3, because expression levels of these genes did not significantly change in insulin-treated cells (Fig. 1C). These data support previous reports (4, 27) indicating that insulin can inhibit the expression of Foxa target genes in hepatocytes, and that the rapid insulin-mediated reduction in transcriptional activity is not caused by alterations in expression levels of Foxa-1, -2, and -3.
The demonstration that insulin signaling leads to a PI3-kinase-mediated decrease in Foxa-2 activity led us to investigate whether downstream targets of PI3-kinase are involved in the modulation of Foxa-2 activity. One prominent downstream target of PI3-kinase, which has been shown to modulate target gene activity by phosphorylation, is Akt (12). To assess the involvement of Akt in mediating the insulin-dependent decrease in Foxa-2 activity we cotransfected HepG2 cells with expression vectors Foxa-1 or Foxa-2 and expression vectors for Akt1/2 or an inactive form of Akt (Akt1K179A). Cotransfection of Foxa-2 with Akt1 or Akt2 completely abolished Foxa-2 activity, whereas expression of the inactive AktK179A protein had no effect (Fig. 1D and data not shown).
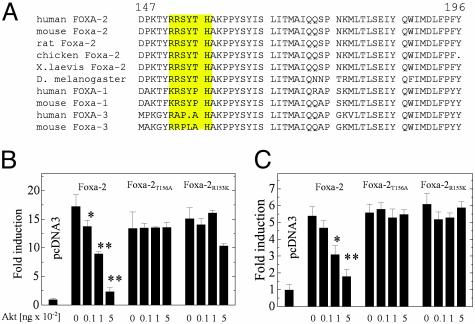
Because our results suggested that Akt could modulate Foxa-2 activity we analyzed the Foxa-2 primary structure for potential Akt phosphorylation sites (28). We identified a putative Akt tyrosine phosphorylation site (RRSYTH) in the human Foxa-2 protein at amino acid position 152–157 that was completely conserved between human, mouse, rat, chicken, and Drosophila melanogaster. No Akt phosphorylation consensus sequences were detected in either Foxa-1 or Foxa-3 (Fig. 2A).
Fig. 2.
Akt regulates Foxa-2 via a conserved Akt phosphorylation site. (A) Sequence alignment of orthologous and paralogous members of the Foxa family. (B) HepG2 cells were transfected with expression vectors for Foxa-2, Foxa-2T156A, or Foxa-2R153K together with Akt in varying concentrations. p6xCdx-TkLuc was used as reporter gene. (C) HepG2 cells were transfected with expression vectors for Foxa-2, Foxa-2T156A, or Foxa-2R153K together with Akt at indicated concentrations. pPEPCK-Luc was used as a reporter gene. In all experiments luciferase activity was normalized to β-galactosidase activity. All vectors were transfected at 125 ng, unless stated otherwise. Values are mean of six independent experiments ± SD. *, P ≤ 0.01; **, P ≤ 0.001.
We next examined whether the identified site was indeed responsible for the Akt-mediated regulation of Foxa-2 phosphorylation. Two different mutants of Foxa-2 were generated: Foxa-2T156A, which cannot be phosphorylated, and Foxa-2R153K, a mutant that is unable to bind to Akt (28). HepG2 cells were cotransfected with p6xCdx-TkLuc, expression vectors for either WT Foxa-2, Foxa-2T156A, or Foxa-2R153K, together with pCMV-HA-Akt-1 or -2. A dose-dependent inhibition of Foxa-2 activity was observed when transfected with increasing amounts of the Akt1 or Akt2 expression vectors (Fig. 2B and data not shown). This decrease was not observed in cotransfections with either Foxa-2T156A or Foxa-2R153K, suggesting that the identified site is indeed responsible for Akt-mediated regulation of Foxa-2 activity. We also analyzed the effect of WT and mutant Foxa-2 proteins on the activity of PEPCK gene transcription by coexpressing these proteins together with pPEPCK-Luc. Transcriptional activity of the PEPCK promoter was increased ≈6-fold in cells cotransfected with Foxa-2. A dose-dependent decrease was observed when increasing amounts of Akt1 or Akt2 expression vectors were cotransfected. No decrease in activity was observed in transfection with either Foxa-2T156A or Foxa-2R153K with increasing amounts of Akt (Fig. 2C).
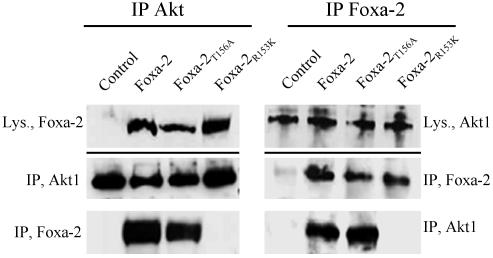
Akt Interacts with and Phosphorylates Foxa-2 at Position T156. To demonstrate that Akt modulates Foxa-2 activity by direct interaction with the putative Akt phosphorylation site we performed immunocoprecipitation experiments. HEK293 cells were transfected with Foxa-2, Foxa-2T156A, or Foxa-2R153K and HA-Akt. Foxa-2 was precipitated by using a polyclonal anti-Foxa-2 antibody, HA-Akt was precipitated with a monoclonal anti-HA antibody. The precipitates were separated by SDS/PAGE and analyzed by Western blotting. As can be seen in Fig. 3, precipitation of HA-Akt led to coprecipitation of Foxa-2 and Foxa-2T156A but not of Foxa-2R153K. Comparison of Foxa-2 and Foxa-2T156A showed that interaction of Foxa-2 with Akt is ≈2-fold weaker than WT Foxa-2. Conversely, precipitation of Foxa-2 yielded similar results, as Akt could be coprecipitated together with Foxa-2 and Foxa-2T156A but not with Foxa-2R153K. No differences in interaction of Foxa-2 and Foxa-2T156A could be seen with this approach (Fig. 3). In addition, by performing in vivo immunoprecipitations in primary hepatocytes, we were able to pull down endogenous Foxa-2 with an anti-Akt1 antibody (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org).
Fig. 3.
Akt interacts with Foxa-2 via the conserved Akt phosphorylation site. HEK293 cells were transfected with expression vectors for Foxa-2, Foxa-2T156A, or Foxa-2R153K together with HA-Akt (5 μg). HA-Akt was precipitated by using anti-HA-antibody (Left). Foxa-2 was precipitated by using an anti-Foxa-2 antibody (Right). Cell lysates (Lys.) and precipitates (IP) were separated by SDS/PAGE and analyzed for Foxa-2 or Akt by Western blotting.
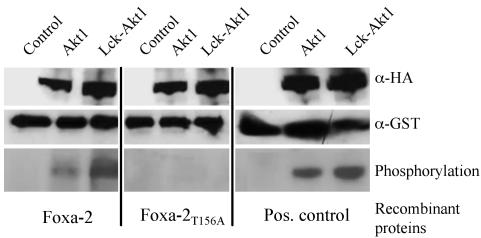
We next tested whether Foxa-2 can be phosphorylated by Akt kinase activity. Recombinant GST-Foxa-2 and GST-Foxa-2T156A proteins were expressed in E. coli BL21 cells, and the soluble protein was purified by anion-exchange chromatography and subsequent size exclusion chromatography. Active Akt was purified from transfected HEK293 cells by immunoprecipitation with anti-HA-antibody. GST-Foxa-2, GST-Foxa-2T156A, or GST-crosstide (24) as positive controls were coincubated with either control cell precipitates (untransfected cells), precipitated HA-Akt, or precipitated constitutive active Akt in the presence of [γ-32P]ATP. Fig. 4 shows that WT Foxa-2 was phosphorylated by either Akt or Lck-Akt, whereas no phosphorylation could be observed when using Foxa-2T156A protein as substrate. Equal loading of WT Foxa-2 and Foxa-2T156A was demonstrated by Western blotting of the phosphorylation reactions (Fig. 4).
Fig. 4.
Akt can phosphorylate Foxa-2 on residue T156. Recombinant GST-Foxa-2, GST Foxa-2T156A, or GST-crosstide [positive control (24)] were incubated in an in vitro kinase reaction together with precipitated HA-Akt or HA-Lck-Akt from HEK293 cell lysates and 0.5 μCi of radiolabeled [γ-32P]ATP (Top and Middle). Untransfected HEK293 cell lysates served as control. Proteins were separated by SDS/PAGE, and phosphorylated proteins were visualized by autoradiography (Bottom). Equal loading was determined by Western blotting using an anti-Foxa-2, anti-GST, or anti-HA antibody.
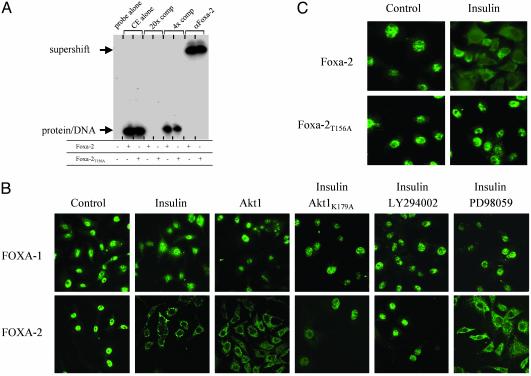
Foxa-2 Phosphorylation by Insulin–PI3-Kinase–Akt Signaling Leads to Nuclear Export. We examined the mechanisms underlying the inhibitory effect of Foxa-2 phosphorylation on the transcriptional activation of target genes. Mechanisms that may account for the inhibitory effects of Akt on Foxa-2 function include an Akt-induced reduction of total Foxa-2 expression levels, impairment of binding to DNA, impairment of Foxa-2's intrinsic transcriptional activation or repressor function, or changes in Foxa-2's nuclear localization. We found that expression of Akt did not significantly change mRNA or protein expression levels of Foxa-2 in HepG2 cells (data not shown). We also compared the DNA binding activity of nuclear extracts from insulin-stimulated HepG2 cells transfected with either WT or Foxa-2T156A expression vectors. Electrophoretic mobility-shift assays were performed to investigate whether mutant Foxa-2 proteins can bind to a Foxa binding site of the Igfpb-1 promoter (25). We found that WT and phosphorylation-deficient mutant Foxa-2T156A from insulin-stimulated cells bound equally to 32P-labeled oligonucleotide probes that contained the Foxa binding site. These data suggested that the phosphorylation state of Foxa-2 does not lead to impairment in DNA binding (Fig. 5A).
Fig. 5.
Phosphorylation of Foxa-2 by Akt-mediated insulin stimulation does not affect DNA binding but leads to nuclear exclusion of Foxa-2. (A) Cell extracts (CE) from insulin-stimulated HEK293 cells, transfected with Foxa-2 or Foxa-2T156A together with Akt (5 μg), were used in an electrophoretic mobility-shift assay. Foxa-2 binding site of Igfbp-1 (25) was used to shift proteins; a consensus Foxa-2 binding site was used for competition. Supershift was performed by using anti-Foxa-2 antibody. (B) Untransfected and Akt (125 ng)-transfected HepG2 cells treated with 50 nM insulin, LY294002, or 10 μM PD98059, alone or in combination, were decorated with anti-Foxa-1 or anti-Foxa-2 antibodies and visualized with an anti-rabbit IgG-Alexa 480 antibody by using laser scanning microscopy. (C) HepG2 cells were transfected with expression vectors for either HA-Foxa-2 or HA-Foxa-2T156A (100 ng) and treated with 50 nM insulin. Cells were decorated with an anti-HA-antibody and visualized with an anti-rabbit IgG-Alexa 480 antibody by using laser scanning microscopy. Control cells were starved for 10 h; all other experiments were performed in medium containing 10% FCS.
We next tested whether Akt-induced phosporylation of Foxa-2 might have an effect on the subcellular distribution of this transcription factor. HepG2 cells were grown to 60% confluency, and endogenous Foxa-1 and Foxa-2 proteins were visualized by immunofluorescence after staining with anti-Foxa-1 and anti-Foxa-2 antibodies. Cells were either examined in the absence or presence of insulin (50 nM) and/or LY294002 or PD98059, and after transfection with either Akt1 or Akt2 expression vectors (Fig. 5B and data not shown). When the endogenous PI3-kinase–Akt pathway was inhibited [by serum starvation (control) or treatment with LY294002 in the presence of insulin] the endogenous Foxa-2 protein was localized almost exclusively in the nucleus. In contrast, cells in which the PI3-kinase–Akt pathway was activated either by treatment with insulin or overexpression of Akt1/2, Foxa-2 was efficiently excluded from the nucleus and largely detected in the cytoplasm (Fig. 5B). Treatment of cells with MEK inhibitor PD98059 had no effect on insulin-stimulated nuclear exclusion of Foxa-2. The subcellular distribution of Foxa-2 in cells that were stimulated with insulin and expressed a dominant negative form of Akt (AktK179A) were resistant to nuclear exclusion. In contrast to the drastic changes in subcellular localization of Foxa-2 upon stimulation of the PI3-kinase–Akt pathway, Foxa-1 protein was not responsive to the activation of this pathway. The same results were obtained when using a glucose-responsive pancreatic β-cell line (Min6, data not shown). To examine whether the effect of Akt on the subcellular localization of Foxa-2 was caused by the phosphorylation of T156 residue, we expressed WT and mutant Foxa-2T156A HA-tagged protein in HepG2 cells and studied the intracellular distribution of this protein in the presence and absence of insulin. In contrast to the WT Foxa-2 protein, Foxa-2T156A was exclusively localized in the nucleus after activation of the PI3-kinase–Akt pathway with insulin (Fig. 5C). Together, these data suggest that insulin stimulation induces phosphorylation via the endogenous PI3-kinase–Akt pathway of a conserved residue, specific for Foxa-2, and that this site plays a crucial role in sequestering Foxa-2 in the cytoplasm, thereby inhibiting Foxa-2's ability to activate transcription of target genes in the nucleus.
Discussion
In the present study we sought to determine whether Foxa-2, a member of the forkhead/winged-helix family of transcription factors, may contribute to the regulation of gene expression by insulin and provide a target for mediating effects of insulin and Akt on gene expression. Foxa-2 has been shown to bind to sites within the regulatory regions of a number of key metabolic genes, including Pepck, G6pc, and Igfbp-1 that are repressed by active insulin–PI3-kinase–Akt signaling (4). However, the molecular mechanism underlying insulin-mediated regulation of gene transcription is not completely understood. In this article, we identify Foxa-2 as a novel substrate for Akt phosphorylation and provide evidence that Foxa-2 transcriptional activity is inhibited by insulin through activation of the PI3-kinase–Akt pathway. Threonine phosphorylation of Foxa-2 at a single conserved residue leads to sequestration of Foxa-2 protein in the cytoplasm and through this mechanism may contribute to coordinately regulate the expression of metabolic genes and genes that control cellular homeostasis.
The forkhead transcription factors Foxa-1, -2, and -3 are structurally related proteins that regulate the expression of three key gluconeogenic enzymes, Pepck, G6pc, and 6-phosphofructo-2-kinase/fructose-2,6-bisphospatase by binding to specific sites in their promoters as monomers (4, 29–32). Foxa proteins have almost identical DNA binding domains, suggesting redundancy in the transcriptional control of these genes. Support for this notion comes from various genetic studies in which single Foxa genes have been inactivated, resulting in little or no dysregulation of Foxa target genes. For example, mice with liver-specific ablation of Foxa-2 seem to have normal glucose homeostasis and gene expression in the liver (33). In this study we show that the Akt phosphorylation site of Foxa-2 (amino acids 152–157) is highly conserved among Foxa-2 proteins but absent in Foxa-1 and -3, albeit being located in a region that is otherwise highly conserved between Foxa genes. Furthermore, only Foxa-2 but not Foxa-1 is inactivated by insulin by nuclear exclusion, suggesting that Foxa-2 is the principal Foxa protein regulated by insulin and that Foxa-1 and -3 may function as basal regulators of gene transcription. The fully compensated transcriptional regulation of Foxa-target genes Pepck, G6pc, and fructose-1,6-bisphosphatase in starved, hypoinsulinemic Foxa-3–/– animals may therefore be caused by augmented transcriptional activation of Foxa-2 protein.
The Akt phosphorylation site of Foxa-2 is highly evolutionary conserved. Similar sites could be identified in various organisms from Homo sapiens to D. melanogaster (Fig. 2 A and data not shown). Interestingly, all these organisms also express at least one other member of the forkhead/winged helix family that does not contain this conserved Akt-phosphorylation site, similar to either Foxa-1 or Foxa-3 (data not shown). The Akt phosphorylation site of Foxa-2 (RRSYTH) differs from the reported “bona fide” consensus sequence determined for Akt (RXRXX(S/T)X) (28, 34). However, structure/function studies have shown that the N-terminal arginine, missing in the Akt phosphorylation site of Foxa-2, is dispensable for phosphorylation by Akt (35). However, we show that arginine residue 153 is required for Akt/Foxa-2 interaction, whereas the threonine residue at position 156 is necessary only for phosphorylation. This finding is also in agreement with the inability of mutant Foxa-2R153K to bind to Akt (Fig. 3). The regulation of Foxa-2 through phosphorylation by the insulin–PI3-kinase–Akt pathway may also contribute to the diabetes-like syndrome observed in mutant mice lacking the Akt2 gene. Mice deficient in Akt2 develop a diabetes-like syndrome as a results of insulin resistance in the liver and skeletal muscle (19). Interestingly, mutant mice have a profound defect in insulin-dependent suppression in hepatic glucose output, reminiscence of transgenic mice overexpressing the Pepck gene in the liver (36). The molecular defect that leads to increased gluconeogenesis in Akt2–/– mice is incompletely understood because of the inherent difficulties to identify substrates of Akt kinases. Our results suggest that Foxa-2 may contribute to the hepatic insulin resistance in Akt2–/– mice by an inability to phosphorylate Foxa-2 and suppress gene transcription of gluconeogenic enzymes. Another forkhead transcription factor that may be regulated by Akt2 and contribute to the failure to suppress gluconeogenesis in Akt2–/– animals is Foxo-1. This protein is a potent activator of Pepck and G6pc gene expression and can be inhibited by Akt-mediated phosphorylation at three distinct sites (16). Inactivation of this protein by phosphorylation can mediate the insulin signal onto G6pc expression but not onto Pepck expression. In epithelial kidney cells (LLC), which express Foxo-1 but lack Foxa-2, Pepck gene expression is unresponsive to regulation by insulin. In contrast, Pepck expression is repressed by insulin in hepatocytes expressing Foxa-2 and Foxo-1 (37). From this it could be speculated that both factors contribute to the reduction of glucose production by regulating the expression of different enzymes involved in gluconeogenesis.
Foxa-2 is also expressed in pancreatic β cells where it regulates the expression of genes such as Pdx-1, Nkx6.2, Sur-1, and Kir6.2 that are important for β cell function and maintaining β cell differentiation (38, 39). It is well known that preservation of the glucose-responsive state of β cells is lost when the extracellular glucose concentration and insulin secretion are chronically elevated (40). Chronic hyperinsulinemia is also an important feature of insulin resistance and obesity and leads to decompensation of β cell function over time. Because insulin signaling is important for normal pancreatic β cell function it is conceivable that chronic stimulation of the insulin–PI3-kinase–Akt pathway in pancreatic β cells leads to inhibition of Foxa-2 activity and may contribute to reduced expression of Foxa-2-regulated genes and dedifferentiation of the β cell phenotype. Transgenic mice overexpressing constitutive active Akt1 in pancreatic β cells develop islet hypertrophy and have markedly increased pancreatic islet mass (41, 42). These morphological changes are accompanied by mild fasting and fed hypoglycemia together with a significant increase in plasma insulin levels. Furthermore, these animals exhibit an increased glucose tolerance in response to glucose injection. β Cell-specific Foxa-2-deficient mice are severely hypoglycemic and show dysregulated insulin secretion in response to both glucose and amino acids (38). However, not an increase in insulin levels but rather an increase in insulin/glucagon ratio is the presumed cause underlying the severe hypoglycemia observed in these animals. As constitutive activation of Akt would lead to suppression of Foxa-2 activity and Foxa-2-regulated gene expression, it can be speculated that inactivation of Foxa-2 may contribute to the observed effect in animals overexpressing Akt in β cells. However, it should be noted that the growth-related changes of pancreatic islets in transgenic mice overexpressing Akt may be triggered by Akt-mediated pathways independent of Foxa-2. Furthermore, β cell function may be impaired in these mice but compensated by increased islet mass.
Genetic studies have demonstrated that the insulin–PI3-kinase–Akt pathway plays pivotal roles in cellular differentiation, proliferation, and metabolism. Studies in C. elegans have demonstrated that specific members of the forkhead transcription factors (e.g., daf-16) may be major targets of insulin receptor signaling downstream of Akt. Mutations of the insulin/Igf-receptor homologue (daf-2), the catalytic subunit of PI3-kinase (age-1), or akt (akt1 and akt2) in C. elegans results in increased longevity and reduced metabolic activity or dauer formation (13–15). Tissue-specific ablation of the insulin receptor in white adipose tissue in the mouse has recently been shown to result in increased longevity, suggesting that basic physiological responses of this pathway may be evolutionary conserved (43). Future studies, using mutant mice, will reveal the role that downstream targets of Akt, such as Foxa-2 or Foxo-1, play in longevity and insulin-regulated target gene expression.
A comprehensive understanding of the signaling pathways that mediate the effects of insulin on the expression of genes known to be important for the regulation of hepatic glucose production and pancreatic β cell function has significant clinical implications. Unrestrained hepatic glucose production and progressive β cell failure are hallmarks of diabetes mellitus. Based on our finding that Foxa-2 is a novel substrate for insulin-mediated Akt regulation we speculate that this pathway may contribute to unrestrained hepatic gluconeogenesis in type 2 diabetes. The design of specific drugs that can inhibit Foxa-2 phosphorylation may be an effective therapeutic approach to the treatment of some forms of diabetes mellitus.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant RO1 DK55033 (to M.S.), the Deutsche Forschungsgemeinschaft (C.W.), the Human Science Frontier Organization (M.S.), a Cancer Research Grant (to D.B.), and an unrestricted grant from Bristol-Myers Squibb (to M.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PEPCK, phosphoenolpyruvate carboxykinase; PI3-kinase, phosphatidylinositol 3-kinase; G6pc, glucose-6-phosphatase; Igfbp-1, insulin growth factor binding protein 1; HA, hemagglutinin; HEK, human embryonic kidney; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase.
See commentary on page 11198.
References
- 1.Kaestner, K. H., Hiemisch, H., Luckow, B. & Schutz, G. (1994) Genomics 20, 377–385. [DOI] [PubMed] [Google Scholar]
- 2.Brennan, R. G. (1993) Cell 74, 773–776. [DOI] [PubMed] [Google Scholar]
- 3.Gerrish, K., Gannon, M., Shih, D., Henderson, E., Stoffel, M., Wright, C. V. & Stein, R. (2000) J. Biol. Chem. 275, 3485–3492. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien, R. M., Noisin, E. L., Suwanichkul, A., Yamasaki, T., Lucas, P. C., Wang, J. C., Powell, D. R. & Granner, D. K. (1995) Mol. Cell. Biol. 15, 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shih, D. Q., Navas, M. A., Kuwajima, S., Duncan, S. A. & Stoffel, M. (1999) Proc. Natl. Acad. Sci. USA 96, 10152–10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granner, D., Andreone, T., Sasaki, K. & Beale, E. (1983) Nature 305, 549–551. [DOI] [PubMed] [Google Scholar]
- 7.Franke, T. F., Yang, S. I., Chan, T. O., Datta, K., Kazlauskas, A., Morrison, D. K., Kaplan, D. R. & Tsichlis, P. N. (1995) Cell 81, 727–736. [DOI] [PubMed] [Google Scholar]
- 8.Franke, T. F., Kaplan, D. R., Cantley, L. C. & Toker, A. (1997) Science 275, 665–668. [DOI] [PubMed] [Google Scholar]
- 9.Hardt, S. E. & Sadoshima, J. (2002) Circ. Res. 90, 1055–1063. [DOI] [PubMed] [Google Scholar]
- 10.Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M. & Hemmings, B. A. (1995) Nature 378, 785–789. [DOI] [PubMed] [Google Scholar]
- 11.Datta, S. R., Dudek, H., Tao, X., Masters, S., Fu, H., Gotoh, Y. & Greenberg, M. E. (1997) Cell 91, 231–241. [DOI] [PubMed] [Google Scholar]
- 12.Datta, S. R., Brunet, A. & Greenberg, M. E. (1999) Genes Dev. 13, 2905–2927. [DOI] [PubMed] [Google Scholar]
- 13.Kenyon, C., Chang, J., Gensch, E., Rudner, A. & Tabtiang, R. (1993) Nature 366, 461–464. [DOI] [PubMed] [Google Scholar]
- 14.Gottlieb, S. & Ruvkun, G. (1994) Genetics 137, 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogg, S., Paradis, S., Gottlieb, S., Patterson, G. I., Lee, L., Tissenbaum, H. A. & Ruvkun, G. (1997) Nature 389, 994–999. [DOI] [PubMed] [Google Scholar]
- 16.Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., Anderson, M. J., Arden, K. C., Blenis, J. & Greenberg, M. E. (1999) Cell 96, 857–868. [DOI] [PubMed] [Google Scholar]
- 17.Kops, G. J., de Ruiter, N. D., De Vries-Smits, A. M., Powell, D. R., Bos, J. L. & Burgering, B. M. (1999) Nature 398, 630–634. [DOI] [PubMed] [Google Scholar]
- 18.Nakae, J., Kitamura, T., Silver, D. L. & Accili, D. (2001) J. Clin. Invest. 108, 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho, H., Mu, J., Kim, J. K., Thorvaldsen, J. L., Chu, Q., Crenshaw, E. B., III, Kaestner, K. H., Bartolomei, M. S., Shulman, G. I. & Birnbaum, M. J. (2001) Science 292, 1728–1731. [DOI] [PubMed] [Google Scholar]
- 20.Cho, H., Thorvaldsen, J. L., Chu, Q., Feng, F. & Birnbaum, M. J. (2001) J. Biol. Chem. 276, 38349–38352. [DOI] [PubMed] [Google Scholar]
- 21.Andjelkovic, M., Jakubowicz, T., Cron, P., Ming, X. F., Han, J. W. & Hemmings, B. A. (1996) Proc. Natl. Acad. Sci. USA 93, 5699–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andjelkovic, M., Alessi, D. R., Meier, R., Fernandez, A., Lamb, N. J., Frech, M., Cron, P., Cohen, P., Lucocq, J. M. & Hemmings, B. A. (1997) J. Biol. Chem. 272, 31515–31524. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz i Altaba, A., Prezioso, V. R., Darnell, J. E. & Jessell, T. M. (1993) Mech. Dev. 44, 91–108. [DOI] [PubMed] [Google Scholar]
- 24.Vandromme, M., Rochat, A., Meier, R., Carnac, G., Besser, D., Hemmings, B. A., Fernandez, A. & Lamb, N. J. (2001) J. Biol. Chem. 276, 8173–8179. [DOI] [PubMed] [Google Scholar]
- 25.Allander, S. V., Durham, S. K., Scheimann, A. O., Wasserman, R. M., Suwanichkul, A. & Powell, D. R. (1997) Endocrinology 138, 4291–4300. [DOI] [PubMed] [Google Scholar]
- 26.Ye, H., Kelly, T. F., Samadani, U., Lim, L., Rubio, S., Overdier, D. G., Roebuck, K. A. & Costa, R. H. (1997) Mol. Cell. Biol. 17, 1626–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotani, K., Ogawa, W., Hino, Y., Kitamura, T., Ueno, H., Sano, W., Sutherland, C., Granner, D. K. & Kasuga, M. (1999) J. Biol. Chem. 274, 21305–21312. [DOI] [PubMed] [Google Scholar]
- 28.Alessi, D. R., Caudwell, F. B., Andjelkovic, M., Hemmings, B. A. & Cohen, P. (1996) FEBS Lett. 399, 333–338. [DOI] [PubMed] [Google Scholar]
- 29.Lemaigre, F. P., Durviaux, S. M. & Rousseau, G. G. (1993) J. Biol. Chem. 268, 19896–19905. [PubMed] [Google Scholar]
- 30.Streeper, R. S., Svitek, C. A., Goldman, J. K. & O'Brien, R. M. (2000) J. Biol. Chem. 275, 12108–12118. [DOI] [PubMed] [Google Scholar]
- 31.Dabeva, M. D., Hurston, E. & Sharitz, D. A. (1995) Am. J. Pathol. 147, 1633–1648. [PMC free article] [PubMed] [Google Scholar]
- 32.Kaestner, K. H., Hiemisch, H. & Schutz, G. (1998) Mol. Cell. Biol. 18, 4245–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sund, N. J., Ang, S. L., Sackett, S. D., Shen, W., Daigle, N., Magnuson, M. A. & Kaestner, K. H. (2000) Mol. Cell. Biol. 20, 5175–5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker, K. S., Deak, M., Paterson, A., Hudson, K., Cohen, P. & Alessi, D. R. (1998) Biochem. J. 331, 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obata, T., Yaffe, M. B., Leparc, G. G., Piro, E. T., Maegawa, H., Kashiwagi, A., Kikkawa, R. & Cantley, L. C. (2000) J. Biol. Chem. 275, 36108–36115. [DOI] [PubMed] [Google Scholar]
- 36.Valera, A., Pujol, A., Pelegrin, M. & Bosch, F. (1994) Proc. Natl. Acad. Sci. USA 91, 9151–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barthel, A., Schmoll, D., Kruger, K. D., Bahrenberg, G., Walther, R., Roth, R. A. & Joost, H. G. (2001) Biochem. Biophys. Res. Commun. 285, 897–902. [DOI] [PubMed] [Google Scholar]
- 38.Sund, N. J., Vatamaniuk, M. Z., Casey, M., Ang, S. L., Magnuson, M. A., Stoffers, D. A., Matschinsky, F. M. & Kaestner, K. H. (2001) Genes Dev. 15, 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, C. S., Sund, N. J., Vatamaniuk, M. Z., Matschinsky, F. M., Stoffers, D. A. & Kaestner, K. H. (2002) Diabetes 51, 2546–2551. [DOI] [PubMed] [Google Scholar]
- 40.Schuit, F., Flamez, D., De Vos, A. & Pipeleers, D. (2002) Diabetes 51, Suppl. 3, S326–S332. [DOI] [PubMed] [Google Scholar]
- 41.Tuttle, R. L., Gill, N. S., Pugh, W., Lee, J. P., Koeberlein, B., Furth, E. E., Polonsky, K. S., Naji, A. & Birnbaum, M. J. (2001) Nat. Med. 7, 1133–1137. [DOI] [PubMed] [Google Scholar]
- 42.Bernal-Mizrachi, E., Wen, W., Stahlhut, S., Welling, C. M. & Permutt, M. A. (2001) J. Clin. Invest. 108, 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bluher, M., Kahn, B. B. & Kahn, C. R. (2003) Science 299, 572–574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.