Abstract
BCR-ABL expression led to a dramatic up-regulation of the IL-3, IL-5, and granulocyte–macrophage colony-stimulating factor receptor β common (IL-3Rβc) and IL-3 receptor β (IL-3Rβ) chains in murine embryonic stem cell-derived hematopoietic cells coincident with an expansion of multipotent progenitors and myeloid elements. This up-regulation required BCR-ABL tyrosine kinase activity and led to IL-3Rβc/β chain tyrosine phosphorylation in the absence of detectable IL-3 production. These results suggested that cytokine-independent IL-3 receptor activation could be a dominant signaling component in BCR-ABL-induced leukemogenesis. To unambiguously define the significance of IL-3 receptor-dependent signaling in BCR-ABL-induced leukemogenesis, BCR-ABL-transduced bone marrow cells deficient in either IL-3Rβc chain or both IL-3Rβc/β chain expression were examined for their ability in generating myeloproliferative disease (MPD). These BCR-ABL-expressing knockout cells were capable of generating MPD similar to control cells, demonstrating that IL-3 receptor activation is not essential for BCR-ABL-induced MPD. However, the IL-3Rβc/β chain could act as a cofactor in BCR-ABL-induced leukemogenesis by activation of its many known oncogenic signaling pathways.
Cytokine signaling has been shown to regulate the growth and development of hematopoietic cell populations (reviewed in ref. 1). Constitutive activation of cytokine receptors has been shown to generate hematopoietic malignancies in animal models (reviewed in ref. 2) and be causative factors in cancers in humans (reviewed in ref. 3).
The Philadelphia (Ph) chromosome generates the BCR-ABL oncogene, which accounts for >90% of chronic myelogenous leukemia (CML) cases and 5–15% of acute B cell lymphoblastic leukemias (reviewed in ref. 4). BCR-ABL tyrosine kinase activity has been demonstrated to be essential for the induction of in vitro cellular transformation (5) and in vivo leukemogenesis (6, 7). Suppression of BCR-ABL tyrosine kinase activity with the small molecule inhibitor imatinib mesylate has been shown to correlate with hematological remission in CML patients (8, 9). However, a significant percentage of CML patients at the acute phases of the disease relapse from imatinib mesylate treatment (10) because of mutations in BCR-ABL rendering the protein resistant to drug inhibition (11, 12).
CML is a hematopoietic stem cell (HSC) disease. The Ph chromosome is initially formed in the HSC and subsequently transduced to all hematopoietic lineages (reviewed in ref. 13). Although the Ph chromosome is present in all hematopoietic lineages, BCR-ABL expression mainly expands progenitors and mature myeloid cells, while causing mild anemia in chronic-phase CML patients (14). The selectivity of BCR-ABL in affecting specific hematopoietic populations suggests that its expression may regulate transcription factors, cytokines, and their receptors that selectively affect the development of these cell populations. The cytokines IL-3 (7, 15–18), vascular endothelial growth factor (VEGF) (19, 20), granulocyte colony-stimulating factor (G-CSF) (16), and granulocyte–macrophage colony-stimulating factor (GM-CSF) (7) have been shown to be up-regulated in BCR-ABL-expressing cells.
An unanswered issue in BCR-ABL signaling is whether any cytokine pathways are essential or play dominant roles in BCR-ABL-induced leukemogenesis. Juvenile myelomonocytic leukemia is a myeloproliferative disease (MPD) that shares some clinical features with CML (reviewed in ref. 21). Data from cultured human juvenile myelomonocytic leukemia cells and murine models implicate deregulated GM-CSF signaling as playing a central role in initiating and maintaining this leukemia (22–25). Based on these data, we reasoned that activated growth factor receptors might cooperate with BCR-ABL in leukemogenesis.
IL-3 is a prominent cytokine shown to be up-regulated on BCR-ABL expression in both hematopoietic cell lines and primary leukemic cells (7, 15–18). IL-3 has been shown to be secreted in the BCR-ABL-expressing FDCP-1 cell line (15), and culture medium from BCR-ABL-expressing embryoid bodies was capable of supporting the growth of IL-3-dependent cell lines 32D and BaF3 (17). A BCR-ABL-induced MPD demonstrated expansion of both BCR-ABL-positive and -negative myeloid cells concomitant with IL-3 transcription (7). These results suggest that BCR-ABL-expressing cells may have a trans effect on expanding normal cells by means of IL-3 secretion.
Although BCR-ABL expression led to growth factor independence in the BaF3 cell line, no detectable amounts of IL-3 were secreted, and these BCR-ABL-expressing cells were incapable of supporting the growth of control BaF3 cells (26). Further, CD34+ CML progenitors mixed in with normal CD34+ cells led to the sole outgrowth of leukemic cells (27). These findings demonstrated predominantly cell-autonomous and not trans effects of BCR-ABL expression on IL-3 up-regulation.
Surprisingly, IL-3 knockout and control bone marrow cells were shown to be similarly competent in generating an acute BCR-ABL-induced MPD in wild-type or IL-3 knockout recipients (28). These results demonstrated that IL-3 was not required in either a cell-autonomous or non-cell-autonomous manner in the generation of BCR-ABL-induced MPD. IL-3Rβc chains have been shown to be activated and capable of inducing cytokine-independent cell growth in the absence of ligand binding by formation of stable interlocking homodimers (29, 30). These results reveal a potential mechanism by which BCR-ABL could activate IL-3 receptor signaling in the absence of ligand production to generate leukemia.
Using an embryonic stem cell (ES) in vitro differentiation system with tetracycline-regulated BCR-ABL expression, we observed a dramatic up-regulation of the IL-3Rβc and IL-3Rβ chains in ES-derived hematopoietic progenitors on BCR-ABL induction. BCR-ABL has been shown to interact with and induce tyrosine phosphorylation of the IL-3Rβc chain (31). Expression of activated forms of IL-3Rβc chains in bone marrow cells generated MPDs (32), and IL-3Rβc chain activation has been shown to activate multiple signaling molecules (reviewed in ref. 33) known to be prominent components in BCR-ABL-induced cellular transformation (reviewed in ref. 34). These combined findings suggested that increased IL-3Rβc/β chain expression could be a dominant mechanism by which BCR-ABL expands hematopoietic progenitors and myeloid elements in a ligand-independent cell-autonomous manner, leading to leukemogenesis.
To define in an unambiguous manner the significance of IL-3 receptor activation in BCR-ABL-induced leukemogenesis, BCR-ABL-transduced bone marrow cells deficient in either IL-3Rβc chain or both IL-3Rβc/β chain expression were examined for their ability to generate MPD. BCR-ABL-expressing IL-3Rβc chain single-knockout or IL-3Rβc/β chain double-knockout bone marrow cells generated MPDs in transplanted recipients similar to control cells. These findings reveal that IL-3 signal transduction is not essential for BCR-ABL-induced MPD.
Materials and Methods
Cell Culture and Induction of MPD. ES in vitro differentiation, induction of BCR-ABL (P210 form) internal ribosome entry site (IRES) enhanced green fluorescent protein (EGFP) expression, and drug inhibition of BCR-ABL tyrosine kinase were as described (35–37).
Wild-type C57BL/6 mice were originally purchased from The Jackson Laboratory, and mice with either a homozygous null mutation of the IL-3Rβc chain (38, 39) or homozygous null mutations in both the IL-3Rβc and IL-3Rβ chains (40) were backcrossed at least 12 generations into a C57BL/6 background and maintained at the University of California, Los Angeles.
Retroviral transduction of BCR-ABL (P210 form) IRES EGFP into wild-type and IL-3Rβc chain-deficient bone marrow cells and transplantation into recipient mice were as described (6, 7). Retroviral infection of IL-3Rβc/β double-knockout bone marrow cells and control cells were performed with the cytokine combination of 25 ng/ml murine thrombopoietin and 100 ng/ml murine FLT-3 ligand instead of IL-3 and IL-6, while maintaining 100 ng/ml murine stem cell factor (SCF) (R & D Systems) because of the inability of IL-3Rβc/β-deficient cells to respond to IL-3 activation (40). This alternative cytokine combination has been shown to stimulate 5-fluorouracil (5-FU)-treated bone marrow cells into cycle and enable retrovirus-mediated gene transfer to HSCs/progenitors (41).
Flow cytometry, pathological evaluation, cell sorting, and cell morphological analyses were as described with the additional phycoerythrin-conjugated anti-IL-3Rβc/β (Pharmingen) antibody used in analyses according to the manufacturer's protocol (36).
RNA Isolation and RT-PCR. Total RNA extraction and RT-PCR were performed as described (36) by using primers as described (42–46). IL-3Rβ amplification was performed with sense 5′-TAATCAACATGACCCTCCTC-3′ and antisense 5′-GCCGAGTGCGCACACGGGCA-3′ IL-3Rβ primers, which crossed exon boundaries and generated a 460-bp cDNA fragment on PCR amplification at 30 cycles with 30-s denaturation at 95°C, 30-s annealing at 55°C, and 60-s primer extension at 72°C. PCRs on cDNAs were initially performed with β-actin primers at 12, 14, and 16 cycles, during the linear range of β-actin amplification, and cDNAs that generated matching levels of β-actin were subsequently used in further PCRs.
Protein Analyses. Immunoblotting was performed as described (36) with the addition of the anti-IL-3Rβc/β chain antibody and the anti-IL-3Rα chain antibody (sc-678 and sc-681, Santa Cruz Biotechnology), which were used according to the manufacturer's protocol. To detect IL-3Rβc/β chain tyrosine phosphorylation, murine IL-3 (R & D Systems) was added on days 5, 6, and 7 of ES differentiation. Hematopoietic progenitors were harvested on day 7 of ES differentiation 10 min after IL-3 addition. To maximally inhibit protein phosphotyrosyl-phosphatases, activated 200 mM NaVO4, prepared as described (47), was combined with 200 mM H2O2 and added to cultures at a 1:1,000 dilution 15 min before cell harvest. One milligram of protein lysates was immunoprecipitated with 2 μg of anti-IL-3Rβc/β chain antibody (sc-678) overnight at 4°C and subsequently bound to protein A-Sepharose beads (Amersham Pharmacia) for 45 min. Bound beads were washed three times with extraction buffer lacking SDS and boiled in SDS-containing sample buffer for 5 min to release immunoprecipitated proteins. Of immunoprecipitated protein lysate, 250 μg equivalents was analyzed for antibody detection.
Results
BCR-ABL Expression Up-Regulates both IL-3Rβc and IL-3Rβ Chain RNA Expression. We have described an ES in vitro differentiation system whereby acute BCR-ABL induction by means of tetracycline regulation led to the selective expansion of ES-derived multipotent and myeloid progenitors with a reduction in erythroblast development (36). We hypothesized that the effects of BCR-ABL could be mediated by regulation of transcription factors, cytokines, and their receptors critical in the generation of these cell types.
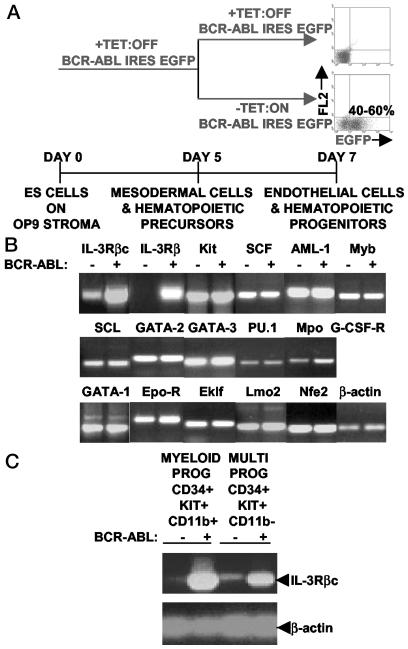
We turned BCR-ABL IRES EGFP expression on at the onset of ES in vitro differentiation into hematopoiesis and harvested EGFP-expressing and control cells 48 h later. These cells were largely composed of multipotent and myeloid progenitors and erythroblasts as determined by fluorescence-activated cell sorting (FACS) (Fig. 1A). BCR-ABL expression led to a 3- to 4-fold expansion in ES-derived multipotent and myeloid progenitors with a 4-fold reduction in erythroblast development (ref. 36; data not shown).
Fig. 1.
BCR-ABL expression up-regulates IL-3Rβc/β mRNA levels in ES-derived multipotent and myeloid progenitors. (A) BCR-ABL expression was turned on at day 5 of ES in vitro cell differentiation, and its effects were analyzed on day 7 in nonadherant hematopoietic progenitors by gating and/or sorting for EGFP-positive cells from cultures without tetracycline (TET). Control cells were obtained from +TET cultures. (B) Of a set of 17 different transcription factors, cytokines and their receptors shown to play critical roles in HSC/progenitor, myeloid, and erythroid cell development, the IL-3Rβc/β chains were the only mRNAs up-regulated on BCR-ABL induction in ES-derived hematopoietic progenitors. Similar results were obtained in three sets of independent experiments. (C) BCR-ABL expression elevated IL-3Rβc mRNA levels in ES-derived multipotent and myeloid progenitors. EGFP-positive and -negative multipotent and myeloid progenitors were FACS sorted to >95% purity, and, in two independent experiments, similar RT-PCR results were obtained. MULTI, multipotent; PROG, progenitors.
RNA was extracted from these cells and RT-PCRs were performed on BCR-ABL + and –samples by using a select group of primers specific for cytokines, their receptors, and hematopoietic transcription factors. The genes examined have been shown to be critical for HSC/progenitor, myeloid, and erythroid cell development through genetic loss-of-function studies (reviewed in ref. 1).
Of a set of 17 different gene primer pairs, the IL-3Rβ and IL-3Rβc chains were the only ones up-regulated on BCR-ABL induction (Fig. 1B). To determine whether this up-regulation occurred in multipotent and myeloid progenitor populations shown to be expanded on BCR-ABL induction (ref. 36; data not shown), these specific cell populations were FACS sorted to >95% purity and analyzed for IL-3Rβc chain RNA expression. Cell surface receptor combinations of CD34+, c-KIT+, and CD11b– were used to define multipotent progenitors, and myeloid progenitors were defined as expressing CD34, c-KIT, and CD11b. BCR-ABL induction elevated IL-3Rβc chain mRNA levels in both multipotent and myeloid lineage-committed progenitors (Fig. 1C).
BCR-ABL Expression Up-Regulates Cell Surface Expression of IL-3Rβc/β Chains. Acute 48-h BCR-ABL induction up-regulated IL-3Rβc/β chain cell surface expression in approximately a third of EGFP-expressing ES-derived hematopoietic progenitors, whereas 3% of control cells had demonstrable expression of these receptors (Fig. 2A). The IL-3Rβc and IL-3Rβ chains are >95% indentical at the amino acid level, and available antibodies against these receptors recognized both receptors and could not distinguish between them. BCR-ABL has been shown to augment cellular adhesion to fibronectin in a tyrosine kinase-independent manner (48). Up-regulation of IL-3Rβc/β chains was inhibited by >90% on addition of the BCR-ABL tyrosine kinase inhibitor imatinib mesylate (Gleevec, STI571, or CP57148B) (Fig. 2A). This cell surface up-regulation of the IL-3Rβc/β chains was similarly distributed among multipotent and myeloid progenitors correlating well with its enhanced mRNA levels (Figs. 1C and 2B).
Fig. 2.
BCR-ABL induces cell surface expression of IL-3Rβc/β in ES-derived hematopoietic cells. (A) BCR-ABL tyrosine kinase activity is required to up-regulate IL-3Rβc/β chain expression in ES-derived hematopoietic progenitors. Imatinib mesylate was added to BCR-ABL cultures at final concentrations of 1 and 10 μM with similar results in triplicate wells. (B) BCR-ABL up-regulates IL-3Rβc/β chain expression in both multipotent and myeloid ES-derived progenitors as defined by cell surface marker analysis. (C) Nine-day induction of BCR-ABL leads to 60–70% of ES-derived hematopoietic cells expressing IL-3Rβc/β chains coincident with low levels of CD11b expression. PE, phycoerythrin.
After 9 days of induction, BCR-ABL further augmented cell surface up-regulation of IL-3Rβc/β chains to between 60% and 70% of ES-derived hematopoietic cells coincident with low levels of CD11b expression (Fig. 2C). This finding is in contrast to control cells, where the large majority of cells expressed a high level of CD11b and lacked IL-3Rβc/β chain expression (Fig. 2C). The maturation status of myeloid cells has been shown to be directly correlated with the level of CD11b expression, where low levels of CD11b expression indicate immature cells and high levels of CD11b expression represent mature cells (49). Long-term BCR-ABL expression was shown to delay myeloid cell maturation during differentiation of FDCP-mix cells (50). IL-3Rβc/β chain up-regulation may play a role in delaying the maturation of myeloid cells observed in BCR-ABL-expressing hematopoietic cells.
BCR-ABL Expression Leads to IL-3Rβc/β Chain Tyrosine Phosphorylation. IL-3 signal transduction can be mediated by cytokine-dependent or growth factor-independent mechanisms (Fig. 3A). The IL-3Rβc chain has been shown to heterodimerize with IL-3Rα, IL-5α, or GM-CSFα receptors on ligand binding, leading to activation of those respective signaling pathways (reviewed in ref. 33). In the mouse, unlike in humans, an additional IL-3Rβ form is expressed that has been shown to exclusively activate IL-3 signal transduction by means of IL-3 and IL-3Rα binding (reviewed in ref. 33). IL-3Rβc chains have been shown to form stable interlocking homodimers in the absence of ligand binding (30), leading to cytokine-independent cell proliferation (29).
Fig. 3.
BCR-ABL up-regulates and tyrosine phosphorylates IL-3Rβc/β chains independent of IL-3 expression. (A) IL-3-dependent and cytokine-independent IL-3Rβc/β chains activated signaling pathways (reviewed in ref. 33). PROL., proliferation; ANTI-APOP., antiapoptosis; DIFF., differentiation; STIM, stimulation. (B) IL-3Rα protein is not up-regulated in ES-derived hematopoietic progenitors on BCR-ABL induction. (C) IL-3 mRNA expression cannot be detected in ES-derived hematopoietic progenitors in the presence or absence of BCR-ABL induction by RT-PCR analyses. WEHI-3 RNA was used as a control for IL-3 RT-PCR. Three sets of BCR-ABL-expressing and control ES-derived hematopoietic progenitors gave similar results. (D) BCR-ABL leads to IL-3Rβc/β chain up-regulation and activation independent of IL-3 production. BaF3 and ES-derived hematopoietic progenitors were stimulated with 0.1, 1.0, and 10 ng/ml IL-3 for 10 min as positive controls with similar results, and NIH 3T3 cells were used as a negative control for IL-3Rβc/β chain expression and tyrosine phosphorylation. NIH, NIH 3T3.
Although the IL-3Rα protein was already expressed in ES-derived hematopoietic progenitors, it was not up-regulated on BCR-ABL induction (Fig. 3B). IL-3 mRNA was not detected in ES-derived hematopoietic progenitors with or without BCR-ABL induction (Fig. 3C). OP9 stromal cells that support the growth and differentiation of ES also did not express IL-3 mRNA (Fig. 3C). Sera used to support ES in vitro differentiation were of bovine origin, suggesting that IL-3 signaling could not have been activated in this murine cell system by this source. These results demonstrate that BCR-ABL expression does not generate all the IL-3-signaling components required for ligand-dependent activation of IL-3Rβc/β chains in this ES in vitro differentiation system.
In the absence of IL-3 stimulus, BCR-ABL expression led to IL-3Rβc/β chain tyrosine phosphorylation at levels similar to ligand stimulation in ES-derived hematopoietic progenitors (Fig. 3D). IL-3 stimulation in non-BCR-ABL-expressing, ES-derived progenitors led to a slight 1.5-fold increase in cell expansion coincident with up-regulation and tyrosine phosphorylation of IL-3Rβc/β chains (Fig. 3D; data not shown). Addition of IL-3 did not augment BCR-ABL-induced expansion of this cell population (data not shown). These results suggest that IL-3-independent activation of IL-3Rβc/β chains could be a dominant signaling mechanism by which BCR-ABL induces leukemia.
IL-3Rβc Chain Expression Is Not Required for BCR-ABL-Induced MPD. To critically determine the role of IL-3Rβc chain up-regulation in BCR-ABL-induced leukemogenesis, IL-3Rβc chain knockout and wild-type bone marrow cells were compared in their ability to generate BCR-ABL-induced MPD. IL-3Rβc chain-deficient mice are reported to have a largely intact hematopoietic compartment with normal survival of neutrophils and macrophages (38, 51) and are unlikely to enhance or suppress myelopoiesis in the absence of BCR-ABL expression.
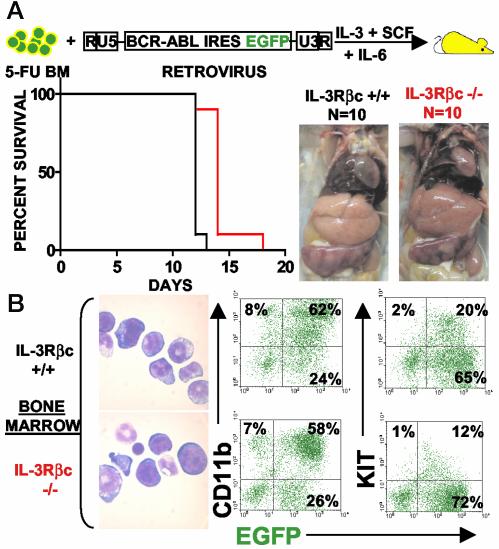
Mice transplanted with BCR-ABL-transduced wild-type or IL-3Rβc chain knockout bone marrow cells developed fatal MPDs (Fig. 4). Nine of 10 mice containing wild-type infected cells generated a deadly MPD 12 days after transplantation and 9 of 10 mice transferred with knockout infected cells developed similarly fatal MPDs 2 days later (Fig. 4A). By the log rank test, the 2-day delay in disease latency between wild-type and IL-3Rβc knockout groups was statistically significant with a P value of <0.0001.
Fig. 4.
IL-3Rβc chain expression is dispensable for BCR-ABL-induced MPD. (A Left) Survival curves of mice transplanted with BCR-ABL-expressing IL-3Rβc knockout or wild-type cells. (Right) Splenomegaly and leukemic cell infiltration in the liver. (B) Cell morphology (Left) and cell surface marker expression of CD11b and KIT in leukemic IL-3Rβc knockout and wild-type cells (Right) appear indistinguishable. Slides were photographed at ×1,000. Cytospins and FACS plots represent an animal from each group, and similar morphologies and percentages of EGFP, CD11b, and KIT expression were obtained in all examined leukemic animals. BM, bone marrow cells.
Both groups of mice developed MPDs with indistinguishable phenotypes characterized by a domination of BCR-ABL-expressing progenitors and myeloid elements as determined by KIT and CD11b expression and cellular morphology (Fig. 4B). Both groups of mice developed similar elevated white blood cell (WBC) counts with an average of 8.35 × 107 ± 1.1 WBCs per ml for both groups of mice and developed similar levels of splenomegaly with average spleen weights of 399 ± 42 mg and 380 ± 60 mg for wild-type and knockout transplanted cells, respectively. Both groups of mice developed leukemic cell infiltration in the liver and pulmonary hemorrhage (Fig. 4A Right and data not shown). These results demonstrate that IL-3Rβc chain expression is not an obligate requirement of BCR-ABL-induced MPD, but it may play a role in accentuating disease progression.
Both IL-3Rβc and IL-3Rβ Chains Are Dispensable for BCR-ABL-Induced MPD. Although IL-3Rβc chain knockout mice lack one of the major receptors required for IL-3 signaling, they express the IL-3Rβ chain whose activation can lead to IL-3 signal transduction (38, 51). We have shown that BCR-ABL induction in ES-derived hematopoietic progenitors led to the up-regulation of both IL-3Rβc and IL-3Rβ chains (Fig. 1B). IL-3Rβc and IL-3Rβ chain double-knockout mice have been shown to maintain normal steady-state hematopoiesis with their bone marrow cells being unresponsive to the cytokines IL-3, IL-5, and GM-CSF (40). To unambiguously determine the requirement of IL-3 signaling in BCR-ABL-induced leukemogenesis, we examined the ability of these double-knockout cells in generating BCR-ABL-induced MPD.
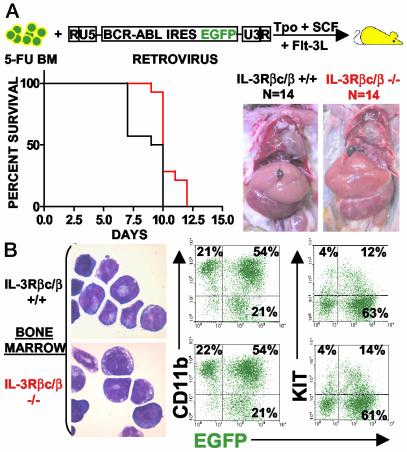
BCR-ABL-transduced IL-3Rβc/β chain double-knockout cells generated MPDs in transplanted mice similar to wild-type BCR-ABL-expressing cells (Fig. 5). All mice transferred with wild-type BCR-ABL-expressing cells from two combined experiments with a total group size of 14 mice developed fatal MPDs by 10 days after transplantation (Fig. 5A Left). In the same combined two experiments, 100% of mice transferred with BCR-ABL-expressing IL-3Rβc/β chain double-knockout cells generated MPDs 12 days after transplantation (Fig. 5A Left). A logrank test comparing survival curves of mice transplanted with wild-type and double-knockout cells gave a P value of 0.0027, suggesting that the difference in disease latencies between the two groups of mice is statistically significant.
Fig. 5.
IL-3Rβc/β chain expression is dispensable for BCR-ABL-induced MPD. (A Left) Survival curves of mice transplanted with BCR-ABL-expressing IL-3Rβc/β double-knockout or wild-type cells. Two experiments, each with seven mice transplanted with BCR-ABL-expressing IL-3Rβc/β double-knockout or wild-type cells, were compiled. (Right) Splenomegaly and leukemic cell infiltration in the liver. (B) Cell morphology (Left) and cell surface marker expression of CD11b and KIT in leukemic IL-3Rβc/β double-knockout and wild-type cells (Right) appear indistinguishable. Cytospins and FACS plots represent an animal from each group, and similar morphologies and percentages of EGFP, CD11b, and KIT expression were obtained in all examined leukemic animals. BM, bone marrow cells.
Leukemic cells in both groups of mice were largely composed of BCR-ABL-expressing CD11b-low, CD11b-high, and KIT-positive cells with morphological features of hematopoietic progenitors, myeloblasts, and mature myeloid elements (Fig. 5B). Both groups of mice had similar elevated WBC counts with an average of 8.62 × 107 ± 3.5 WBCs per ml and 11.65 × 107 ± 8.1 WBCs per ml for wild-type and double-knockout transplanted cells, respectively, and developed similar levels of splenomegaly with spleens weighing 447 ± 25 mg and 515 ± 50 mg for wild-type and double-knockout transplanted cells, respectively. Both groups of mice developed leukemic cell infiltration in the liver and pulmonary hemorrhage (Fig. 5A Right; data not shown).
These results demonstrate that cell-autonomous activation of IL-3 signaling is not essential for BCR-ABL-induced MPD but may contribute to BCR-ABL oncogenesis.
Discussion
IL-3Rβc/β Chains and BCR-ABL Activate Redundant Signaling Pathways. IL-3Rβc/β chains have been shown to interact with SHC and GRB2 adaptors, leading to RAS-MAPK activation (reviewed in ref. 33). The same protein adaptors have been demonstrated to associate with BCR-ABL, correlating with stimulation of the RAS-MAPK pathway in fibroblast cell lines that do not express IL-3 receptors (reviewed in ref. 34). IL-3Rβc/β chains and BCR-ABL can both up-regulate MYC expression (reviewed in refs. 33 and 34). Expression of dominant-negative forms of RAS and MYC have demonstrated that these molecules are essential for BCR-ABL cellular transformation (reviewed in ref. 34). However, the capability of BCR-ABL to up-regulate these molecules independent of IL-3 activation may account for the dispensability in IL-3Rβc/β chain expression in generating BCR-ABL-induced MPD. Alternatively, activated receptors for other cytokines such as Kit, Flt3, and/or G-CSFR might cooperate with BCR-ABL and thereby compensate for inactivation of IL-3Rβc/β chains.
Up-Regulation of IL-3Rβc/β Chains May Enhance BCR-ABL-Induced Leukemogenesis. In addition to up-regulating RAS and MYC, IL-3Rβc/β chain activation has been shown to turn on the PI3K/AKT pathway (reviewed in ref. 33). RAS, MYC, and PI3K/AKT activation has been shown to generate and/or contribute to the development of cancers in humans (reviewed in ref. 52). Elevated levels of IL-3Rα in acute myeloid leukemia have been associated with enhanced blast proliferation and poor prognosis (53). These findings suggest that BCR-ABL up-regulation of IL-3Rβc/β chains may lead to the development of more aggressive leukemias because of IL-3Rβc/β chain activation of oncogenic signaling pathways.
IL-3Rβc/β Chain Up-Regulation May Contribute to CML Pathogenesis by Deregulating Hematopoietic Progenitor Maturation. Although IL-3Rβc/β chain double-knockout bone marrow cells were capable of generating BCR-ABL-induced acute 7- to 12-day MPDs, inspection of the phenotype of these leukemic cells revealed that they expressed high levels of CD11b and low-to-undetectable levels of IL-3Rβc/β chains (data not shown), indicative of mature myeloid cells. This finding is in contrast to the effect of BCR-ABL expression in ES-derived hematopoietic progenitors during the same time frame, which suppressed myeloid cell maturation and led to the expansion of immature myeloid elements characterized by IL-3Rβc/β chain up-regulation and low levels of CD11b expression. These findings demonstrate that IL-3Rβc/β chains are dispensable for the generation of BCR-ABL-induced acute MPD initiating from mature myeloid cells, but suggest that IL-3Rβc/β chain up-regulation may play a role in the evolution of CML originating from HSCs/progenitors. IL-3Rβc/β chain expression in HSCs/progenitors may delay progenitor cell maturation, which could lead to an increase in the number of immature myeloid progenitors that we have described (36). This, in turn, could lead to the overproduction of mature myeloid cells seen in chronic phase CML.
IL-3 Receptor Activation Could Contribute to the Persistence of Ph+ HSCs in CML Patients. High concentrations of IL-3 have been shown to promote the self-renewal of a murine multipotential hematopoietic cell line (54), and expression of a V449E-activated form of the IL-3Rβc chain has been shown to generate a murine MPD dominated by blasts with the surface phenotype of HSCs as defined by KITlow, Thy-1.2low, Sca-1+, and lineage-negative expression (32). BCR-ABL transduced IL-3-deficient cells have been shown to be growth factor-independent for up to 2 weeks in culture and generate an acute 15- to 20-day BCR-ABL-induced MPD (28, 45). However, the same cells are incapable of long-term cytokine-independent growth, and prolonged in vitro expansion of these cells abrogated its ability to generate a murine multilineage leukemia unlike similarly treated wild-type cells (45). These results suggest that IL-3 receptor activation could be a potential mechanism by which BCR-ABL-expressing HSCs/progenitors are self-renewed to continually perpetuate disease development.
Targeted inactivation of the IL-3 signal transduction pathway by small molecule inhibitors that suppress IL-3 receptor tyrosine kinase activity or antagonist antibodies, such as BION-1, that block cytokine activation of IL-3 receptors (55) may prevent self-renewal of Ph+ HSCs/progenitors. Such treatment may lead to the eventual exhaustion of Ph+ HSCs/progenitors, a target population that current imatinib mesylate therapy cannot eliminate.
Acknowledgments
We are grateful to Richard Murray for providing the IL-3Rβ knockout mice; Andrew Kim for breeding and 5-FU treating IL-3Rβ knockout mice; James Johnson for excellent technical assistance with animal work; Shirley Quan for coordinating transfers of various mouse strains; Fiona Willis for technical expertise on the FACSVantage cell sorter; and Barbara Anderson for excellent preparation of the manuscript. This work was supported, in part, by National Institutes of Health Grants CA72614 and CA76204, the National Health and Medical Research Council of Australia, and The Sylvia and Charles Viertel Charitable Foundation. O.N.W. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: ES, embryonic stem cell; Ph, Philadelphia; CML, chronic myelogenous leukemia; HSC, hematopoietic stem cell; GM-CSF, granulocyte–macrophage colony-stimulating factor; IRES, internal ribosome entry site; EGFP, enhanced GFP; SCF, stem cell factor; MPD, myeloproliferative disease; 5-FU, 5-fluorouracil; FACS, fluorescence-activated cell sorting.
References
- 1.Zhu, J. & Emerson, S. G. (2002) Oncogene 21, 3295–3313. [DOI] [PubMed] [Google Scholar]
- 2.Kelly, L. M., Liu, Q., Kutok, J. L., Williams, I. R., Boulton, C. L. & Gilliland, D. G. (2002) Blood 99, 310–318. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, W. S. & Nicola, N. A. (1998) Leuk. Res. 22, 1097–1111. [DOI] [PubMed] [Google Scholar]
- 4.Deininger, M. W., Goldman, J. M. & Melo, J. V. (2000) Blood 96, 3343–3356. [PubMed] [Google Scholar]
- 5.Pendergast, A. M., Gishizky, M. L., Havlik, M. H. & Witte, O. N. (1993) Mol. Cell. Biol. 13, 1728–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pear, W. S., Miller, J. P., Xu, L., Pui, J. C., Soffer, B., Quackenbush, R. C., Pendergast, A. M., Bronson, R., Aster, J. C., Scott, M. L. & Baltimore, D. (1998) Blood 92, 3780–3792. [PubMed] [Google Scholar]
- 7.Zhang, X. & Ren, R. (1998) Blood 92, 3829–3840. [PubMed] [Google Scholar]
- 8.Druker, B. J., Talpaz, M., Resta, D. J., Peng, B., Buchdunger, E., Ford, J. M., Lydon, N. B., Kantarjian, H., Capdeville, R., Ohno-Jones, S. & Sawyers, C. L. (2001) N. Engl. J. Med. 344, 1031–1037. [DOI] [PubMed] [Google Scholar]
- 9.Kantarjian, H., Sawyers, C., Hochhaus, A., Guilhot, F., Schiffer, C., Gambacorti-Passerini, C., Niederwieser, D., Resta, D., Capdeville, R., Zoellner, U., et al. (2002) N. Engl. J. Med. 346, 645–652. [DOI] [PubMed] [Google Scholar]
- 10.Druker, B. J., Sawyers, C. L., Kantarjian, H., Resta, D. J., Reese, S. F., Ford, J. M., Capdeville, R. & Talpaz, M. (2001) N. Engl. J. Med. 344, 1038–1042. [DOI] [PubMed] [Google Scholar]
- 11.Gorre, M. E., Mohammed, M., Ellwood, K., Hsu, N., Paquette, R., Rao, P. N. & Sawyers, C. L. (2001) Science 293, 876–880. [DOI] [PubMed] [Google Scholar]
- 12.Shah, N. P., Nicoll, J. M., Nagar, B., Gorre, M. E., Paquette, R. L., Kuriyan, J. & Sawyers, C. L. (2002) Cancer Cell 2, 117–125. [DOI] [PubMed] [Google Scholar]
- 13.Kabarowski, J. H. S. & Witte, O. N. (2000) Stem Cells 18, 399–408. [DOI] [PubMed] [Google Scholar]
- 14.Lichtman, M. A. (1995) in Williams Hematology, eds. Buetler, E., Lichtman, M. A., Coller, B. S. & Kipps, T. J. (McGraw–Hill, New York), pp. 298–324.
- 15.Hariharan, I. K., Adams, J. M. & Cory, S. (1988) Oncol. Res. 3, 387–399. [PubMed] [Google Scholar]
- 16.Jiang, X., Lopez, A., Holyoake, T., Eaves, A. & Eaves, C. (1999) Proc. Natl. Acad. Sci. USA 96, 12804–12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters, D. G., Klucher, K. M., Perlingeiro, R. C., Dessain, S. K., Koh, E. Y. & Daley, G. Q. (2001) Oncogene 20, 2636–2646. [DOI] [PubMed] [Google Scholar]
- 18.Holyoake, T. L., Jiang, X., Jorgensen, H. G., Graham, S., Alcorn, M. J., Laird, C., Eaves, A. C. & Eaves, C. J. (2001) Blood 97, 720–728. [DOI] [PubMed] [Google Scholar]
- 19.Mayerhofer, M., Valent, P., Sperr, W. R., Griffin, J. D. & Sillaber, C. (2002) Blood 100, 3767–3775. [DOI] [PubMed] [Google Scholar]
- 20.Aguayo, A., Kantarjian, H., Manshouri, T., Gidel, C., Estey, E., Thomas, D., Koller, C., Estrov, Z., O'Brien, S., Keating, M., et al. (2000) Blood 96, 2240–2245. [PubMed] [Google Scholar]
- 21.Emanuel, P. D., Shannon, K. M. & Castleberry, R. P. (1996) Mol. Med. Today 2, 468–475. [DOI] [PubMed] [Google Scholar]
- 22.Emanuel, P. D., Bates, L. J., Castleberry, R. P., Gualtieri, R. J. & Zuckerman, K. S. (1991) Blood 77, 925–929. [PubMed] [Google Scholar]
- 23.Bollag, G., Clapp, D. W., Shih, S., Adler, F., Zhang, Y. Y., Thompson, P., Lange, B. J., Freedman, M. H., McCormick, F., Jacks, T. & Shannon, K. (1996) Nat. Genet. 12, 144–148. [DOI] [PubMed] [Google Scholar]
- 24.Largaespada, D. A., Brannan, C. I., Jenkins, N. A. & Copeland, N. G. (1996) Nat. Genet. 12, 137–143. [DOI] [PubMed] [Google Scholar]
- 25.Birnbaum, R. A., O'Marcaigh, A., Wardak, Z., Zhang, Y. Y., Dranoff, G., Jacks, T., Clapp, D. W. & Shannon, K. M. (2000) Mol. Cell 5, 189–195. [DOI] [PubMed] [Google Scholar]
- 26.Daley, G. Q. & Baltimore, D. (1988) Proc. Natl. Acad. Sci. USA 85, 9312–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, S., Haylock, D. N., Levesque, J. P., McDiarmid, L. A., Samels, L. M., To, L. B., Simmons, P. J. & Hughes, T. P. (1998) Blood 92, 2461–2470. [PubMed] [Google Scholar]
- 28.Li, S., Gillessen, S., Tomasson, M. H., Dranoff, G., Gilliland, D. G. & Etten, R. A. V. (2001) Blood 97, 1442–1450. [DOI] [PubMed] [Google Scholar]
- 29.Patel, N., Herrman, J. M., Timans, J. C. & Kastelein, R. A. (1996) J. Biol. Chem. 271, 30386–30391. [DOI] [PubMed] [Google Scholar]
- 30.Carr, P. D., Gustin, S. E., Church, A. P., Murphy, J. M., Ford, S. C., Mann, D. A., Woltring, D. M., Walker, I., Ollis, D. L. & Young, I. G. (2001) Cell 104, 291–300. [DOI] [PubMed] [Google Scholar]
- 31.Wilson-Rawls, J., Xie, S., Liu, J., Laneuville, P. & Arlinghaus, R. B. (1996) Cancer Res. 56, 3426–3430. [PubMed] [Google Scholar]
- 32.McCormack, M. P. & Gonda, T. J. (1999) Oncogene 18, 7109–7199. [DOI] [PubMed] [Google Scholar]
- 33.Guthridge, M. A., Stomski, F. C., Thomas, D., Woodcock, J. M., Bagley, C. J., Berndt, M. C. & Lopez, A. F. (1998) Stem Cells 16, 301–313. [DOI] [PubMed] [Google Scholar]
- 34.Maru, Y. (2001) Int. J. Hematol. 73, 308–322. [DOI] [PubMed] [Google Scholar]
- 35.Era, T. & Witte, O. N. (2000) Proc. Natl. Acad. Sci. USA 97, 1737–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong, S., McLaughlin, J., Cheng, D. & Witte, O. N. (2003) Blood 101, 4088–4097. [DOI] [PubMed] [Google Scholar]
- 37.Era, T., Wong, S. & Witte, O. N. (2001) Methods Mol. Biol. 185, 83–95. [DOI] [PubMed] [Google Scholar]
- 38.Robb, L., Drinkwater, C. C., Metcalf, D., Li, R., Kontgen, F., Nicola, N. A. & Begley, C. G. (1995) Proc. Natl. Acad. Sci. USA 92, 9565–9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishinakamura, R., Miyajima, A., Mee, P. J., Tybulewicz, V. L. & Murray, R. (1996) Blood 88, 2458–2464. [PubMed] [Google Scholar]
- 40.Scott, C. L., Robb, L., Papaevangeliou, B., Mansfield, R., Nicola, N. A. & Begley, C. G. (2000) Blood 96, 1588–1590. [PubMed] [Google Scholar]
- 41.Wognum, A. W., Visser, T. P., Peters, K., Bierhuizen, M. F. & Wagemaker, G. (2000) Hum. Gene Ther. 11, 2129–2141. [DOI] [PubMed] [Google Scholar]
- 42.Keller, G., Kennedy, M., Papayannopoulou, T. & Wiles, M. V. (1993) Mol. Cell. Biol. 13, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delassus, S., Titley, I. & Enver, T. (1999) Blood 94, 1495–1503. [PubMed] [Google Scholar]
- 44.Vannucchi, A. M., Linari, S., Lin, C. S., Koury, M. J., Bondurant, M. C. & Migliaccio, A. R. (1999) J. Cell. Physiol. 180, 390–401. [DOI] [PubMed] [Google Scholar]
- 45.Jiang, X., Ng, E., Yip, C., Eisterer, W., Chalandon, Y., Stuible, M., Eaves, A. & Eaves, C. J. (2002) Blood 100, 3731–3740. [DOI] [PubMed] [Google Scholar]
- 46.Ide, H., Seligson, D. B., Memarzadeh, S., Xin, L., Horvath, S., Dubey, P., Flick, M. B., Kacinski, B. M., Palotie, A. & Witte, O. N. (2002) Proc. Natl. Acad. Sci. USA 99, 14404–14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon, J. A. (1991) Methods Enzymol. 201, 477–482. [DOI] [PubMed] [Google Scholar]
- 48.Wertheim, J. A., Forsythe, K., Druker, B. J., Hammer, D., Boettiger, D. & Pear, W. S. (2002) Blood 99, 4122–4130. [DOI] [PubMed] [Google Scholar]
- 49.Warren, M. K. & Vogel, S. N. (1985) J. Immunol. 134, 982–989. [PubMed] [Google Scholar]
- 50.Pierce, A., Spooncer, E., Wooley, S., Dive, C., Francis, J. M., Miyan, J., Owen-Lynch, P. J., Dexter, T. M. & Whetton, A. D. (2000) Oncogene 19, 5487–5497. [DOI] [PubMed] [Google Scholar]
- 51.Scott, C. L., Hughes, D. A., Cary, D., Nicola, N. A., Begley, C. G. & Robb, L. (1998) Blood 92, 4119–4127. [PubMed] [Google Scholar]
- 52.Perkins, A. S. & Stern, D. F. (1997) in Cancer: Principles and Practice of Oncology, eds. Vincent T. DeVita, V. T., Hellmann, S. & Rosenberg, S. A. (Lippincott–Raven, Philadelphia), pp. 79–102.
- 53.Testa, U., Riccioni, R., Militi, S., Coccia, E., Stellacci, E., Samoggia, P., Latagliata, R., Mariani, G., Rossini, A., Battistini, A., Lo-Coco, F. & Peschle, C. (2002) Blood 100, 2980–2988. [DOI] [PubMed] [Google Scholar]
- 54.Heyworth, C. M., Dexter, T. M., Kan, O. & Whetton, A. D. (1990) Growth Factors 2, 197–211. [DOI] [PubMed] [Google Scholar]
- 55.Sun, Q., Jones, K., McClure, B., Cambareri, B., Zacharakis, B., Iversen, P. O., Stomski, F., Woodcock, J. M., Bagley, C. J., D'Andrea, R. & Lopez, A. F. (1999) Blood 94, 1943–1951. [PubMed] [Google Scholar]