Abstract
Prions are usually quantified by bioassays based on intracerebral inoculation of mice that are slow, imprecise, and costly. We have isolated neuroblastoma N2a sublines highly susceptible to mouse prions, as evidenced by accumulation of infectivity and the scrapie form of prion protein (PrPSc), and developed quantitative in vitro assays for prion infectivity. In the scrapie cell (SC) assay, susceptible N2a cells are exposed to prion-containing samples for 3 days, grown to confluence, and split 1:10 three times, and the proportion of PrPSc-containing cells is determined with automated counting equipment. In a log/log plot, the dose–response is linear over two logs of prion concentrations. The SC assay is about as sensitive as the mouse bioassay, 10 times faster, >2 orders of magnitude less expensive, and suitable for robotization. SC assays performed in a more time-consuming end point titration format extend the sensitivity and show that infectivity titers measured in tissue culture and in the mouse are similar.
In prion diseases the normal host prion protein (PrPC) is converted to a largely protease-resistant, aggregated form designated PrPSc (1). PrPSc is a surrogate marker for prion disease, but its level does not necessarily correlate with the infectivity titer (2–5). Infectivity is usually quantified by injecting samples intracerebrally (i.c.) into indicator animals, usually mice, and determining the time to appearance of definitive clinical symptoms (incubation time method) (6) or by injecting serial dilutions of the sample and determining the dilution at which 50% of the animals acquire scrapie (endpoint method) (7).
Several cell lines can be infected by prions, as evidenced by the accumulation of PrPSc and/or infectivity after multiple passages (8–14). Murine neuroblastoma-derived N2a cells are susceptible to certain strains of mouse prions, such as the mouse-adapted Rocky Mountain Laboratory (RML) scrapie strain (15); however, only a small proportion of cells accumulate detectable levels of PrPSc. Selected sublines of N2a cells are more susceptible to the RML strain than the original stock but nonetheless resistant to murine strains such as Me7, 301V, 22A, and others (16–19). The suitability of such N2a sublines for an in vitro infectivity assay was assessed by exposing them to serial dilutions of RML strain-infected mouse brain homogenate and determining the dilution at which a blotting assay for PrPSc became negative. The assay was reported to be “several orders of magnitude” less sensitive than the mouse bioassay (16).
We have isolated N2a sublines highly susceptible to RML scrapie prions and those that are relatively resistant. We observed that infected monolayers, when blotted onto membranes and assayed for PrPSc, gave a speckled pattern, suggesting that microcolonies of infected cells, embedded in a lawn of uninfected cells, were being displayed. Indeed, when infected cultures were diluted so that single colonies could be distinguished, some appeared PrPSc positive and others negative. We then succeeded in visualizing individual PrPSc-positive cells filtered onto membranes of an enzyme-linked immunospot (ELISPOT) plate. We established conditions under which the proportion of PrPSc-positive colonies or cells was a function of the prion titer and developed two quantitative assays, the colony assay and the scrapie cell (SC) assay. With the N2aPD88 and N2aPK1 cell lines we could quantify RML prion concentrations about as low as those that can be determined in the mouse bioassay. The colony assay requires no special equipment but is work intensive, whereas the automated SC assay requires special equipment but allows the convenient evaluation of large numbers of samples. The assay can be completed in 2 weeks, as compared with 20 weeks in the most rapid mouse bioassay (20), and is 2 orders of magnitude cheaper. The SC assay can be performed in an endpoint titration format, and although more time consuming, it is more sensitive and robust, capable of detecting infectivity at a 10–8 dilution of RML strain-infected mouse brain, which is beyond the range of the standard mouse bioassay. Infectivity titers measured in tissue culture and in the mouse by the endpoint assays are similar.
Materials and Methods
Preparation of Prion-Infected Brain Homogenates. The mouse prion strains 22A (I858) and ME7 (I857) were propagated in VM and C57BL/6 mice, respectively. Bovine spongiform encephalopathy prions (I218) were propagated in C57BL/6 mice and the hamster strain 263K (I3113) in Syrian hamsters. To prepare RML prion-infected brain homogenates, 10 brains of terminally sick CD1 mice were homogenized in PBS with a glass homogenizer. The 10% (wt/vol) homogenate (designated I2424) was stored at –80°C. All other brain homogenates were obtained from M. Desbrulais, Medical Research Council Prion Unit.
Subcloning, Propagation, and Storage of N2a Cells. N2a cell lines were cultured in Opti-MEM-10% FCS (OFCS) (Invitrogen). The susceptible cell line N2a/Gary was isolated essentially as described (18) from N2a stock obtained from the American Type Culture Collection (N2aATCC), except that the cells were not transfected with a PrP expression plasmid. Clones of N2a/Gary cells were challenged with a 10–6 dilution of RML strain, to yield clone N2aPD88, which gave the highest response in the SC assay. Further subcloning of N2aPD88 yielded the more sensitive N2aPK1 line (data not shown). N2aATCC cells or subclones thereof, such as the N2aR33 and NN2a lines, that failed to accumulate PrPSc after exposure to a 10–4 RML strain dilution were designated nonsusceptible. Chosen clones were expanded for three passages, and 50 aliquots each were frozen at –80°C in OFCS and 6% DMSO and stored in liquid nitrogen. Working stocks are propagated for not more than about four 1:10 splits, because prolonged subculturing leads to diminished susceptibility (R. Kilner and P.-C.K., unpublished observations).
The Colony Assay. Unless stated otherwise, all steps were carried out at room temperature. TBST (10 mM Tris·HCl, pH 8.0/150 mM NaCl/0.1% Tween 20) was freshly prepared from a 10× stock solution. Twenty thousand susceptible N2aPD88 cells (doubling time ≈24 h) in 200 μl of OFCS were plated in each well of a 96-well plate. After 16 h at 37°C the medium was replaced by a 0.3-ml assay sample in OFCS. After 3 days the confluent monolayer (≈105 cells) was suspended by gentle pipetting. A 30-μl aliquot of each sample was placed into a well of a 96-well plate containing 270 μl of OFCS and grown to confluence. The cells were split 1:10 two more times, and after reaching confluence after the third split, aliquots of 103, 104, and 105 cells were deposited onto 25-mm Thermanox coverslips (Nunc, Fisher Scientific) in six-well plates. Uninfected cells were added to make up a total of 105 cells in each case, because the efficiency of the subsequent proteinase K (PK) digestion depends on the amount of protein transferred to the membrane in the next step. After 4 days at 37°C, the cell layer was blotted onto a nitrocellulose membrane (TransBlot transfer medium, nitrocellulose, 0.45 μm, Bio-Rad) soaked in lysis buffer (50 mM Tris·HCl, pH 8.0/150 mM NaCl/0.5% sodium deoxycholate/0.5% Triton X-100). The membrane was dried 1 h at 37°C, incubated in lysis buffer containing 5 μg/ml PK (specific activity 30 millianson units/mg, Merck Pharmaceuticals) for 90 min at 37°C, rinsed twice with water, and incubated in 2 mM PMSF (Sigma) for 10 min. The membrane was placed into 3 M guanidinium thiocyanate (>99%, Sigma-Aldrich), 10 mM Tris·HCl (pH 8.0) for 10 min, rinsed five times with water, incubated in Superblock (Tris buffer saline) blocking buffer (Pierce) for 1 h, shaken with anti-PrP antibody 6H4 (21) (Prionics, Zürich; 1:5,000) or ICSM18 (22) (provided by S. Hawke, Imperial College Faculty of Medicine, London; 1:5,000) in TBST-1% nonfat dry milk (Marvel, Premier Brands UK, Lincs, U.K.) for 1 h, then washed with TBST four times for 5 min. The membrane was incubated with horseradish peroxidase-conjugated anti-mouse IgG1 antibody (Zymed; 1:7,500 in TBST, 1% nonfat dry milk) for 1 h. After washing five times for 5 min with TBST, membranes were dried at room temperature, soaked for 3 min with ECL reagent (Super Signal West Pico, Pierce), and exposed to x-ray film (Hyperfilm ECL, Amersham Pharmacia). PrPSc-positive colonies were counted and expressed relative to the number of plated cells.
The SC Assay. All solutions were filtered. TBST was prepared fresh from a 10× stock solution. Unless stated otherwise, all procedures were at room temperature. Cells were infected and propagated as above until confluent after the third split. The cells were suspended in 250 μl of PBS, and 25,000 or 5,000 cells (see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org, for the cell counting procedure) were placed into each of at least four wells (containing 200 μl PBS) of an ELISPOT plate (activated with 70% ethanol; Multi Screen Immobilon-P 96-well Filtration Plates, sterile, Millipore) and washed twice with PBS by suction. After 1hat50°C, 50 μl of PK (0.5 μg/ml in lysis buffer for 25,000 cells; 0.25 μg/ml for 5,000 cells) was added to each well, incubated 90 min at 37°C, and suctioned off. The PK concentration relative to the number of cells on the membrane is critical for optimal sensitivity and signal-to-noise ratio. The samples were washed with PBS, exposed to 2 mM PMSF for 10 min, and washed again with PBS. After incubation with 160 μl of 3 M guanidinium thiocyanate, 10 mM Tris·HCl (pH 8.0) for 10 min, the filters were washed four times with PBS and incubated with 160 μl of Superblock for 45 min. Fifty microliters of PrP antibody ICSM18 (0.6 μg/ml, in TBST/1% nonfat milk powder) was added; after 1 h the supernatant was suctioned off, and the wells were washed seven times with TBST. Fifty microliters of alkaline phosphatase-conjugated anti-IgG1 (Southern Biotechnology Associates; 1:4,500 in TBST/1% milk powder) was added for 1 h. Wells were washed eight times with TBST, and 50 μl of alkaline phosphatase conjugate substrate (prepared as recommended by Bio-Rad) was added for 8 min (signal-to-noise ratios were higher after incubation for 16 min) and followed by two washes with water. Plates were stored in the dark at –20°C.
PrPSc-positive cells were counted by using the Zeiss KS ELISPOT system (Stemi 2000-C stereo microscope equipped with a Hitachi HV-C20A color camera and a KL 1500 LCD scanner and wellscan software from Imaging Associates, Bicester, U.K.). The settings were optimized to give a maximal ratio of counts for PrPSc-positive samples relative to negative control samples, while keeping the sample counts as high as possible. Parameters were adjusted by using the training feature of the program according to the instructions of the manufacturer.
Serial dilutions of a titered RML standard should be run in parallel to the unknown samples. The dilutions of the standard and the samples should be chosen such that the spot values lie between ≈500 and 1,000; the number of cells plated (25,000 or 5,000) should be the same for sample and standard.
Results
Isolation of N2a-Derived Cell Lines Highly Susceptible or Relatively Resistant to RML Prions. Neuroblastoma N2a sublines cloned from an American Type Culture Collection stock culture were screened for susceptibility to RML prions as described (16, 18). A clone designated N2a/Gary was ≈100-fold more sensitive to RML prions than the original stock. A second round of subcloning yielded N2aPD88, a cell line 1,000-fold more sensitive than noncloned cells (see below). More recently we isolated an even more sensitive cell line, N2aPK1, by subcloning N2aPD88 cells. Several nonsusceptible lines, such as clones NN2a or N2aR33, were isolated by exposure to higher concentrations of RML homogenates and selection for cells showing no PrPSc accumulation. N2aR33 cells were ≈100-fold less sensitive than N2aPD88 cells (Fig. 1a). No significant difference in cell-surface PrPC was found between N2aPD88 cells and the nonsusceptible line NN2a, as assessed by fluorescence-activated cell sorting analysis (data not shown).
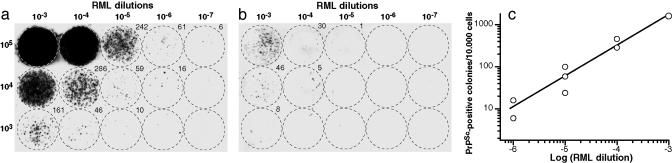
Fig. 1.
Dose–response relationship between the dilution of infected RML homogenate and the number of PrPSc-positive colonies determined by the colony assay. Twenty thousand susceptible N2aPD88 cells (a) and nonsusceptible N2aR33 cells (b) were exposed to the indicated dilutions of RML strain (I2424, see Table 1) in OFCS. After 3 days, the cells were split 1:10 and grown for 3 days. After two more splits, 105, 104, or 103 cells were mixed with noninfected N2aPD88 cells for a total of 105 cells and plated on coverslips. After 4 days, PrPSc-positive colonies were identified as described in Materials and Methods. Where countable, the number of positive colonies is indicated at the upper right of the blot. (c) Double-logarithmic plot of RML strain dilution versus number of PrPSc-positive colonies. The countable positive colonies were recalculated per 10,000 plated cells.
The Colony Assay for Prion Infectivity. Susceptible N2aPD88 cells and nonsusceptible N2aR33 cells were exposed in parallel to serial 10-fold dilutions (10–3 to 10–7; 10–1 = 10% brain homogenate) of an RML prion-infected brain homogenate (108.4 LD50 units per g) and split successively three times 1:10 to dilute out PrPSc particles carried over from the inoculum (see below). After reaching confluence, 105, 104, or 103 cells were grown on coverslips for 4 days and subjected to the cell blot assay (16). Fig. 1a shows that the number of PrPSc-positive N2aPD88 colonies increased with the number of cells plated, although not quite in proportion. In Fig. 1c, colony numbers that were countable and significant (between 10 and 300) plotted against the RML strain dilution on a log-log scale, gave an approximately linear relationship between 10–3 and 10–6 (correlation coefficient = 0.96). Fig. 1 shows that nonsusceptible N2aR33 cells required a 100-fold higher RML strain concentration to give the same number of colonies as N2aPD88 cells.
In this experiment one PrPSc-positive colony per 10,000 cells plated was equivalent to ≈10 LD50 units, when measured at a dilution of 10–5 or 10–6 of the RML homogenate. Using a reference curve such as that of Fig. 1c the infectivity of an unknown sample can be determined.
The SC Assay for Mouse Prion Infectivity. We developed a procedure to visualize single PrPSc-positive cells by an ELISA. Therefore, rather than growing infected cells to colonies, we filtered defined numbers of cells onto the membranes of ELISPOT plates and subjected them to lysis and PK digestion. PrPSc-positive cells were detected by using a monoclonal PrP antibody and alkaline phosphatase-linked anti-IgG1 antiserum, and counted by using the Zeiss KS ELISPOT system (see Fig. 3a). The equipment can resolve up to 1,000–1,500 cells per well; thereafter the response becomes nonlinear (Fig. 5, which is published as supporting information on the PNAS web site).
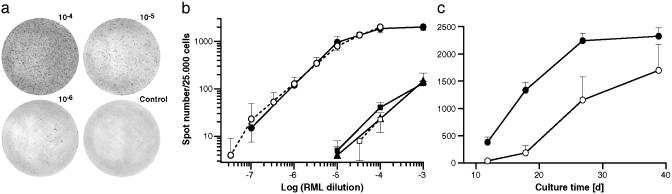
Fig. 3.
Dose- and time-dependent response of susceptible and nonsusceptible N2a cells to various dilutions of RML homogenate in the SC assay. (a) Representative wells of an ELISPOT plate showing spots given by N2aPD88 cells exposed to the indicated dilutions of RML strain or to medium (control), as described in experiment 1. (b) Double-logarithmic plot of spot number versus RML strain dilution. Experiment 1 (filled symbols): 20,000 susceptible cells (N2aPD88, circles) and nonsusceptible cells (N2aR33, squares; N2aATCC, triangles) were incubated for 3 days with serial 1:10 dilutions of RML strain (I2424) in OFCS, as indicated. The cells were split three times 1:10, aliquots of 25,000 cells were filtered onto membranes of an ELISPOT plate, and PrPSc-positive cells were determined by the SC assay. Mean values of quadruplicate measurements ± SD are shown. Background values of noninfected cells (19 ± 4 for N2aPD88, 5 ± 1 for N2aR33, and 9 ± 2 for N2aATCC) were subtracted. N2aPD88 cells are ≈3 logs more susceptible than the nonsusceptible cells. Experiment 2 (open symbols): Serial 1:3.3 dilutions of RML strain (I2424, 10–1) into uninfected CD1 brain homogenate (10–1) were diluted 1:1,000 in OFCS, to give a final dilution of total brain homogenate of 10–4 and the dilution of RML strain indicated. Twenty thousand susceptible (N2aPD88, circles) and nonsusceptible cells (N2aR33, squares) were exposed to the samples and processed as in experiment 1. Mean values of six measurements ± SD are shown. Background values of cells incubated with noninfected CD1 homogenate (16 ± 7 for N2aPD88 and 6 ± 6 for N2aR33) were subtracted. In the double-logarithmic plot the relationship between spot number and RML strain dilution is linear from 10–7 to 10–5 (correlation coefficient r = 0.94). The values for the dilutions 10–7 and 3.3 × 10–7 are significantly over background (P = 0.006 and P = 0.0007, respectively; Mann–Whitney U test) but not that for 3.3 × 10–8.(c) Time-dependent increase in the proportion of infected cells. PD88 cells were exposed to a 10–5 (•) or 10–6 (○) dilution of RML strain (I2424) in OFCS for 3 days and split 1:10 every third day. The spot number per 25,000 cells was determined at the times indicated. Mean values of triplicate measurements and SD are shown. Spot numbers were corrected for background values of noninfected cells (16 ± 7). The rate of accrual is initially ≈20–30% per day but levels off as the saturation value of detectable infected cells is approached (≈2,000 per 25,000 cells).
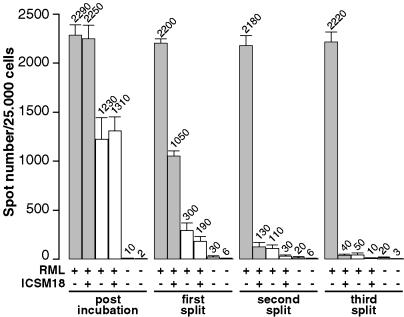
Cells exposed to infected brain homogenates cultures must be split three times 1:10 to dilute out PrPSc-positive particles present in the homogenates, as shown in the experiment of Fig. 2. Susceptible N2aPD88 and nonsusceptible NN2a cells were exposed to RML strain-infected brain homogenate for 3 days, split 1:10, grown to confluence, and split again 1:10. This procedure was repeated once more. At each step, after reaching confluence, the proportion of PrPSc-positive particles was determined by the SC assay. All procedures were carried out in parallel in the presence of monoclonal PrP antibody ICSM18 (22) to prevent accumulation of PrPSc in chronically scrapie-infected N2a cells (18). Three days after infection the number of immunoreactive “spots” was similar whether or not PrP antibodies were present, both in susceptible and nonsusceptible cells (Fig. 2), showing that at this stage the vast majority of spots could be accounted for by PrPSc particles from the inoculum, probably adsorbed to cells. After the third split, the spot number in the antibodytreated samples diminished to background levels, as compared with ≈2,200 spots in the absence of antibody, showing that the original PrPSc particles had been diluted out and/or degraded completely. Similarly, nonsusceptible cells exposed to the RML strain and split three times 1:10 typically gave a reduced number of spots of ≈50, <2.5% of the original level. This value was further reduced to 10 by PrP antibody, suggesting that nonsusceptible cells were also susceptible to infection, albeit at very low efficiency. Thus, the PrPSc-positive spots detected after three 1:10 splits are virtually entirely caused by de novo-infected cells.
Fig. 2.
Clearance of PrPSc particles from the initial inoculum after three successive 1:10 splits of infected cells. Twenty thousand susceptible N2aPD88 cells (gray bars) and nonsusceptible NN2a cells (white bars) were cultured for 16 h, plated in a 96-well plate, and exposed for 3 days to a 10–4 dilution of RML strain (I2424) in OFCS, in the absence (–) or presence (+) of the anti-PrP mAb ICSM 18 (5 μg/ml, added 4 h before infection), as indicated. As controls, N2aPD88 and NN2a cells were incubated with OFCS only. Three days after infection, the cells were suspended, octaplicate aliquots of 25,000 cells were filtered onto the membranes of an ELISPOT plate, and the PrPSc-positive signals were quantified by the SC assay. Both susceptible and nonsusceptible cells gave rise to PrPSc-positive signals. An anti-PrP antibody that abrogates PrPSc accumulation did not diminish the spot number, demonstrating that all spots can be accounted for by PrPSc particles from the inoculum. After three 1:10 splits, nonsusceptible and susceptible cells incubated with antibody showed <2% of the spots given by susceptible cells. The numbers for noninfected N2aPD88 cells and NN2a cells, the fifth and sixth bar of each group, respectively, represent the background. Mean values and SD are shown.
We next determined the dose–response to RML prions. Susceptible N2aPD88 and nonsusceptible N2a cells were exposed to serial dilutions of RML strain-infected brain homogenates (10–3 to 10–7) and processed as above. Fig. 3a depicts magnifications of ELISPOT filters with samples at various dilutions, and Fig. 3b shows that log-log plots from two independent experiments are linear between 10–7 and 10–5 dilutions and that response is dynamic up to a 10–4 dilution. The flattening off of the response is partly caused by the failure in resolving individual PrPSc-positive cells at values exceeding ≈1,000 cells and can be to some extent overcome by screening 5,000 or fewer, rather than 25,000 cells (Fig. 5).
The Ratio of Infected Cells to Total Cells Increases During Propagation. PD88 cells were exposed to a 10–5 or 10–6 dilution of RML strain (I2424) for 3 days and split 1:10 every third day, and the spot number per 25,000 cells was determined at various times up to 39 days (Fig. 3c). In the case of infection with 10–5 RML strain, the proportion of PrPSc-positive cells increased ≈3.5-fold between the third and the fifth split but leveled off as it approached a “saturation” value of ≈2,300 per 25,000 cells, after a total of eight 1:10 splits. Because the background values do not increase, the signal-to-noise ratio for dilute prion samples, and therefore the sensitivity of the assay can be improved several fold by extending the postinfection culture period, albeit at the cost of prolonging the assay time.
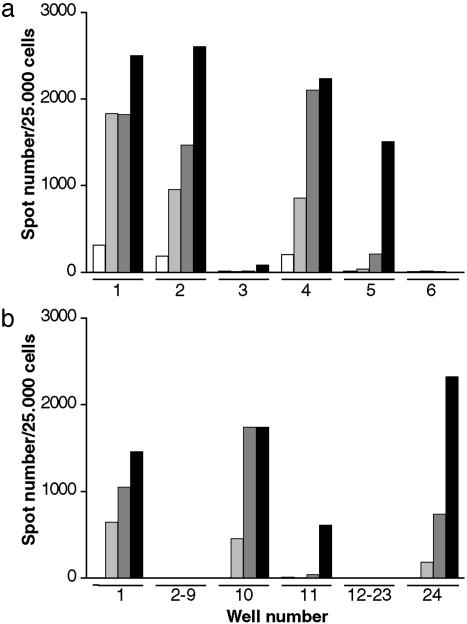
SC Assay in an Endpoint Titration Format. Six wells with susceptible N2a cells (N2aPK1) were exposed to a 10–7 dilution of RML strain (I2424) and 24 wells to a 10–8 dilution for 3 days and then split 1:10 every 3 days. After 3, 5, 8, and 12 splits, 25,000 cells were subjected to the SC assay and scored for the presence of PrPSc-containing cells. As shown in Fig. 4, several wells were already positive after the third split, and the proportion of scrapie-infected cells in those wells increased with successive splits. In the case of the 10–7 dilution 5 of 6, and in the case of the 10–8 dilution 4 of 24 wells were positive. The dilution leading to 50% positive wells is calculated to be 10–7.5 (7), and the brain therefore contains 108 tissue culture LD50 units per g, which is ≈40% of the value 108.4 LD50 units per g determined by the mouse bioassay for the same RML preparation (see Table 2, which is published as supporting information on the PNAS web site). The sensitivity of this assay can be further increased by a modified splitting protocol (Table 2).
Fig. 4.
Serial 1:10 dilutions of RML strain (I2424, 10%) into uninfected brain homogenate (CD1, 10%) were prepared. Twenty thousand susceptible cells (subclone N2aPK1) were incubated for 3 days with 300 μlof10–7 (a) and 10–8 (b) dilution of RML strain in OFCS. The cells were split 3 times 1:10 (12 days, white bars), 5 times (18 days, light gray bars), 8 times (27 days, gray bars), and 12 times (39 days, black bars); 25,000 cells were subjected to the SC assay. Mean ± SD of background values of noninfected cells were 11 ± 6, 16 ± 13, 14 ± 5, and 9 ± 2 at 12, 18, 27, and 39 days, respectively. After 39 days of culture, wells with spot numbers exceeding the mean value of the background by 5 SDs were scored as positive. The dilution leading to 50% positive wells was 10–7.5, as calculated by following ref. 7. The brain therefore contained 107.5/(0.3) = 108 tissue culture LD50 units per g.
Comparison of the Mouse Bioassay and the SC Assay. Brain homogenate (I2424) from terminally ill, RML strain-infected CD1 mice was serially diluted 10-fold, and 30-μl aliquots of each dilution were injected i.c. into each of five mice. In parallel, 100-μl aliquots were administered i.p. Table 1 shows that all mice succumbed to scrapie when injected i.c. with dilutions down to 10–6, but only three of five mice at a 10–7 dilution and none of five mice at a 10–8 dilution. The same homogenate assayed by the SC assay gave 31 ± 21 spots per 25,000 cells (SC units) for the 10–7 dilution and 15 ± 8 for the 10–8 dilution. Both values are significantly above background (P < 0.001) as determined by the Mann–Whitney U test. By these criteria, the mouse infectivity assay and the SC assay have about the same sensitivity.
Table 1. Infectivity assay of RML scrapie-infected brain homogenate.
| Mouse endpoint assay*
|
SC assay†
|
||
|---|---|---|---|
| Dilution‡ | Sick/total | Incubation time, days ± SD | SC units ± SD (background) |
| i.c. inoculation | |||
| 10-2 | 5/5 | 61 ± 3 | Not done |
| 10-4 | 5/5 | 74 ± 5 | 2,030 ± 230 (25 ± 17) |
| 10-5 | 5/5 | 88 ± 9 | 926 ± 226 (6 ± 5) |
| 10-6 | 4/5 | 102 ± 12 | 98 ± 46 (4 ± 4) |
| 10-7 | 3/5 | 116 ± 27 | 31 ± 21 (4 ± 3) |
| 10-8 | 0/5 | >155 | 15 ± 8 (5 ± 3) |
| i.p. inoculation | |||
| 10-2 | 5/5 | 92 ± 3 | — |
| 10-4 | 5/5 | 116 ± 4 | — |
| 10-5 | 3/5 | 123 ± 16 | — |
| 10-6 | 0/5 | >155 | — |
Inoculated mice were monitored every second day and culled after definitive onset of disease. The experiment was terminated at 155 days postinoculation. The dilution leading to 50% scrapie-sick mice after i.c. inoculation was 10-6.9 and for the i.p. inoculation 10-5.2, calculated following Reed and Muench (7). The brain therefore contained 108.4 LD50 units per g as assayed i.c. and 106.7 LD50 units per g as assayed i.p.
RML strain-infected brain homogenates (12424) were diluted in OFCS or 10-4 CD1 homogenate. The values, positive cells per 25,000 plated cells, are averages of 30 independent measurements of each sample on sensitive (N2aPD88) cells (backgrounds not subtracted) and 16 measurements of the backgrounds on nonsusceptible N2aATCC or N2aR33 cells exposed to the same samples. The value for the 10-7 dilution, 31 ± 21 (n = 30), was over a background of 4 ± 3 (n = 16) and differed significantly from it (P < 0.0001, Mann-Whitney U test).
Serial 10-fold dilutions of the 10% (wt/vol) 12424 brain homogenate (10% = 10-1 dilution) were prepared with PBS or PBS containing 0.01% uninfected brain homogenate, so that all samples (except the 10-2 dilution) had a final concentration of 0.01% brain homogenate. Thirty microliters of the dilutions was inoculated i.c. or 100 μl i.p. into Tga20 mice (20). For the SC assay, the 10% homogenate was diluted into OFCS.
Considering that 300 μl of brain homogenate (I2424) at a 10–5 dilution resulted in 926 scrapie-positive cells per 25,000 cells (926 SC units) and at a 10–6 dilution in 98 SC units, there are ≈108.5 SC units per g of brain. The mouse brain from which the same samples were derived contained 108.4 LD50 units per g. Thus, under the conditions of the experiment, one SC unit is equivalent to ≈1.3 LD50 unit [108.5/(108.4)]. However, it should be noted that the SC units are arbitrarily defined and moreover depend on antibody avidity and concentrations and the parameters chosen for counting by the ELISPOT equipment. Therefore, in comparing results from independent assays, it is mandatory to include a reference prion preparation, preferably one that has been titered in the mouse bioassay.
SC assays were performed on four RML brain homogenates with known LD50 titers (Table 3, which is published as supporting information on the PNAS web site). Although the values for SC units per g of brain fell within a narrow range of 3–8 × 108, the ratio of SC units to LD50 units ranged from 0.2 to 3.5 as determined by the mouse bioassay in different laboratories; this spread is likely caused by the imprecision of the mouse bioassay (23).
Infectivities in Homogenates of Mouse Brains at Different Times After Inoculation Determined by the Mouse Bioassay and the SC Assay. To further compare the sensitivity of the mouse infectivity and the SC assay, brain homogenates from uninfected CD1 mice were assayed by both methods at different times after inoculation. As shown in Table 4, which is published as supporting information on the PNAS web site, neither method detected infectivity at days 25 and 41 after i.c. inoculation, but after 61 and 140 days infectivity was detected by both methods. Borderline infectivity found at 19 days by the mouse bioassay, probably caused by residual inoculum, was not reflected in the SC assay.
Exposure of N2aPD88 Cells to Various Prion Strains. Susceptible N2aPD88 and nonsusceptible N2aR33 cells (20,000 per well) were incubated for 3 days with dilutions (5 × 10–3 and 10–3) of (untitered) brain homogenates from terminally sick animals infected with the murine ME7 (I857) or 22A (I858) strain, hamster 263K prions (I3113), bovine spongiform encephalopathy prions (I218, passaged in C57BL/6 WT mice), or variant Creutzfeldt–Jakob disease prions (I335). After five 1:10 splits none of the samples showed a significant increase of PrPSc-positive spots over the values given by nonsusceptible cells (22 ± 9 spots). In contrast, N2aPD88 cells exposed to a 10–4 RML strain yielded 1,500 PrPSc-positive spots after the fifth split.
Discussion
We have isolated cell lines and developed procedures that allow infectivity assays about as sensitive as the mouse bioassay, routinely detecting infectivity down to a 10–7 dilution of brain homogenate from terminally RML strain scrapie sick mice. It should be noted that although the criterion for determining cell infection is the accumulation of protease-resistant PrP, what is in fact being measured by the assay is transmission of infectious agent to the cells. Interestingly, highly and poorly susceptible N2a cells did not differ significantly in their PrP expression level, confirming that even though PrP expression is essential for prion replication, it is not the limiting factor (18).
Because RML strain-infected brain homogenate contains PrPSc particles that attach to the N2a cell monolayer and give signals indistinguishable from infected cells, the cultures must be split three times 1:10 to dilute out these particles. Under the assay conditions we describe PrPSc-positive spots are virtually entirely caused by de novo-infected cells. In the course of cell propagation the proportion of infected cells increases, resulting in an enhanced signal-to-noise ratio and consequently in an increased sensitivity of the assay, albeit at the cost of a longer assay time. The increase in the proportion of infected cells could be caused by cell-to-cell transmission (24) and/or by an increase in the PrPSc level in cells that already were infected but not detectable under our assay conditions. A less likely explanation is that PrPSc-producing cells outgrow noninfected cells.
We describe two approaches to quantify infected cells. The colony assay requires no sophisticated equipment; however, the procedure is work intensive and does not lend itself to automation. The SC assay can be performed in a 96-well plate format and ≈1,000 spots per well can be readily resolved under the microscope and counted automatically by appropriate ELISPOT scanning equipment in a few minutes.
When plotted on a double logarithmic scale, the linear range of the standard SC assay, using N2aPD88 cells, is between 10–7 and 10–5 for brain homogenates from terminal, RML strain-infected mice, whereas the dynamic response extends to 10–4. Sensitivity can be further increased by performing five to eight rather than three splits, because the proportion of infected cells, but not the background, increases with time, resulting in a higher signal-to-noise ratio.
The SC assay can be applied in an endpoint titration format. Dilute prion preparations are distributed into 6–24 wells and subjected to the SC assay after 8–12 splits, and the number of wells containing PrPSc-positive cells are scored. After eight splits, negative wells give a background of 10–20 spots whereas positive wells usually exhibit several hundred or more spots. The tissue culture endpoint assay and the mouse endpoint assay give quite similar LD50 values for the same RML brain homogenate, namely 108 and 108.4, respectively. This good agreement, although of practical interest, may be to some degree accidental. After injection of prions into mouse brain, most infectivity is rapidly lost from the brain (25), by leakage and perhaps partly by inactivation. On the other hand, exposure of cell cultures to a prion suspension may not allow all potentially infectious particles to initiate infection. The sensitivity of the endpoint titration can be further increased by performing the initial splits at a 1:3 ratio every other day (Table 2) and using 48 or 96 wells per sample.
The SC assay is presently restricted in its applicability because our N2a sublines are resistant to bovine prions, even after they have been passaged through the mouse, to human variant Creutzfeldt–Jakob disease, hamster 263K, and murine ME7 and 22A prions.
The standard SC assay, from start to finish, takes 14 days. The mouse infectivity assay, using PrP-overexpressing tga20 mice, takes ≈140 days (20). The problems inherent to quantification of the infectivity assay in the mouse are set forth in ref. 23. The costs of assaying a single prion sample by endpoint titration, with use of 30 mice as in Table 1, are ≈£450, as compared with the expenditure of <£2 per sample (quadruplicate determination including controls) for the SC assay. The SC assay can likely replace a large proportion of the in vivo assays for RML prions and thereby contribute to the well-being of mice worldwide.
Supplementary Material
Acknowledgments
We thank E. McKintosh for help with mouse infectivity assays, A. Vehlow for contributing to the assay development, R. Kilner for technical assistance, M. Desbrulais for brain homogenates, and R. Young for preparing the figures. We are indebted to J. Collinge for generous support and encouragement, A. Aguzzi for valuable suggestions and materials, and P. Schwarz for help with the mouse bioassays. This study was supported by grants from the Medical Research Council (to J. Collinge and C.W.).
Abbreviations: ELISPOT, enzyme-linked immunospot; i.c., intracerebrally; OFCS, Opti-MEM-10% FCS; SC, scrapie cell; PK, proteinase K; PrP, prion protein; RML, Rocky Mountain Laboratory.
References
- 1.Prusiner, S. B. (1991) Science 252, 1515–1522. [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi, S., Katamine, S., Yamanouchi, K., Kishikawa, M., Moriuchi, R., Yasukawa, N., Doi, T. & Miyamoto, T. (1993) J. Gen. Virol. 74, 2117–2123. [DOI] [PubMed] [Google Scholar]
- 3.Shaked, G. M., Fridlander, G., Meiner, Z., Taraboulos, A. & Gabizon, R. (1999) J. Biol. Chem. 274, 17981–17986. [DOI] [PubMed] [Google Scholar]
- 4.Manson, J. C., Jamieson, E., Baybutt, H., Tuzi, N. L., Barron, R., McConnell, I., Somerville, R., Ironside, J., Will, R., Sy, M. S., et al. (1999) EMBO J. 18, 6855–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lasmezas, C. I., Deslys, J. P., Robain, O., Jaegly, A., Beringue, V., Peyrin, J. M., Fournier, J. G., Hauw, J. J., Rossier, J. & Dormont, D. (1997) Science 275, 402–405. [DOI] [PubMed] [Google Scholar]
- 6.Prusiner, S. B., Cochran, S. P., Groth, D. F., Downey, D. E., Bowman, K. A. & Martinez, H. M. (1982) Ann. Neurol. 11, 353–358. [DOI] [PubMed] [Google Scholar]
- 7.Reed, J. & Muench, H. (1938) Am. J. Hyg. 27, 493–497. [Google Scholar]
- 8.Race, R. E., Fadness, L. H. & Chesebro, B. (1987) J. Gen. Virol. 68, 1391–1399. [DOI] [PubMed] [Google Scholar]
- 9.Butler, D. A., Scott, M. R., Bockman, J. M., Borchelt, D. R., Taraboulos, A., Hsiao, K. K., Kingsbury, D. T. & Prusiner, S. B. (1988) J. Virol. 62, 1558–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubenstein, R., Carp, R. I. & Callahan, S. M. (1984) J. Gen. Virol. 65, 2191–2198. [DOI] [PubMed] [Google Scholar]
- 11.Vilette, D., Andreoletti, O., Archer, F., Madelaine, M. F., Vilotte, J. L., Lehmann, S. & Laude, H. (2001) Proc. Natl. Acad. Sci. USA 98, 4055–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Follet, J., Lemaire-Vieille, C., Blanquet-Grossard, F., Podevin-Dimster, V., Lehmann, S., Chauvin, J. P., Decavel, J. P., Varea, R., Grassi, J., Fontes, M. & Cesbron, J. Y. (2002) J. Virol. 76, 2434–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birkett, C. R., Hennion, R. M., Bembridge, D. A., Clarke, M. C., Chree, A., Bruce, M. E. & Bostock, C. J. (2001) EMBO J. 20, 3351–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schatzl, H. M., Laszlo, L., Holtzman, D. M., Tatzelt, J., DeArmond, S. J., Weiner, R. I., Mobley, W. C. & Prusiner, S. B. (1997) J. Virol. 71, 8821–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandler, R. L. (1961) Lancet 1, 1378–1379. [DOI] [PubMed] [Google Scholar]
- 16.Bosque, P. J. & Prusiner, S. B. (2000) J. Virol. 74, 4377–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishida, N., Harris, D. A., Vilette, D., Laude, H., Frobert, Y., Grassi, J., Casanova, D., Milhavet, O. & Lehmann, S. (2000) J. Virol. 74, 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enari, M., Flechsig, E. & Weissmann, C. (2001) Proc. Natl. Acad. Sci. USA 98, 9295–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beranger, F., Mange, A., Solassol, J. & Lehmann, S. (2001) Biochem. Biophys. Res. Commun. 289, 311–316. [DOI] [PubMed] [Google Scholar]
- 20.Fischer, M., Rülicke, T., Raeber, A., Sailer, A., Moser, M., Oesch, B., Brandner, S., Aguzzi, A. & Weissmann, C. (1996) EMBO J. 15, 1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 21.Korth, C., Stierli, B., Streit, P., Moser, M., Schaller, O., Fischer, R., Schulz-Schaeffer, W., Kretzschmar, H., Raeber, A., Braun, U., et al. (1997) Nature 390, 74–77. [DOI] [PubMed] [Google Scholar]
- 22.White, A. R., Enever, P., Tayebi, M., Mushens, R., Linehan, J., Brandner, S., Anstee, D., Collinge, J. & Hawke, S. (2003) Nature 422, 80–83. [DOI] [PubMed] [Google Scholar]
- 23.McLean, A. R. & Bostock, C. J. (2000) Philos. Trans. R. Soc. London B 355, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanu, N., Imokawa, Y., Drechsel, D. N., Williamson, R. A., Birkett, C. R., Bostock, C. J. & Brockes, J. P. (2002) Curr. Biol. 12, 523–530. [DOI] [PubMed] [Google Scholar]
- 25.Büeler, H., Aguzzi, A., Sailer, A., Greiner, R. A., Autenried, P., Aguet, M. & Weissmann, C. (1993) Cell 73, 1339–1347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.