Abstract
The predominant mode of HIV transmission worldwide is via heterosexual contact, with the cervico-vaginal mucosa being the main portal of entry in women. The cervico-vaginal mucosa is naturally colonized with commensal bacteria, primarily lactobacilli. To address the urgent need for female-controlled approaches to block the heterosexual transmission of HIV, we have engineered natural human vaginal isolates of Lactobacillus jensenii to secrete two-domain CD4 (2D CD4) proteins. The secreted 2D CD4 recognized a conformation-dependent anti-CD4 antibody and bound HIV type 1 (HIV-1) gp120, suggesting that the expressed proteins adopted a native conformation. Single-cycle infection assays using HIV-1HxB2 carrying a luciferase reporter gene demonstrated that Lactobacillus-derived 2D CD4 inhibited HIV-1 entry into target cells in a dose-dependent manner. Importantly, coincubation of the engineered bacteria with recombinant HIV-1HxB2 reporter virus led to a significant decrease in virus infectivity of HeLa cells expressing CD4–CXCR4–CCR5. Engineered lactobacilli also caused a modest, but statistically significant, decrease in infectivity of a primary isolate, HIV-1JR-FL. This represents an important first step toward the development of engineered commensal bacteria within the vaginal microflora to inhibit heterosexual transmission of HIV.
The predominant mode of HIV transmission worldwide is via heterosexual contact (1). Women are particularly at risk for HIV infection, because the efficiency of HIV transmission from male to female is greater than for the reverse (2). There are few means by which women can actively protect themselves against HIV infection, particularly in the absence of a protective vaccine. The need to develop new methods of HIV prevention that are controlled by women is urgently recognized by health organizations worldwide.
The cervico-vaginal mucosa is the main site of HIV entry in women. In healthy women of child-bearing age, the protective mucosa in the vagina is populated with microflora typically dominated by lactobacilli and their dominance over pathogenic anaerobes is positively associated with vaginal health (3). Depletion or disturbances of vaginal lactobacillus flora has been associated with establishment of opportunistic infections like bacterial vaginosis and an increased risk of acquiring HIV type 1 (HIV-1) (4). The principal lactobacillus species isolated from the vaginal mucosa of healthy women are Lactobacillus crispatus, Lactobacillus jensenii, and Lactobacillus gasseri (5, 6). These three species are efficient colonizers of the vaginal mucosa and likely exist in a natural “biofilm” composed of bacteria and extracellular matrix materials (7).
The vaginal microflora of women is part of a dynamic ecosystem. Through genetic engineering, the vaginal microflora may be further enhanced to form an efficient protective shield against the transmission of sexually transmitted diseases like HIV. Our approach involves the genetic modification of natural human isolates of lactobacilli to express high-affinity HIV-binding proteins. These proteins can be associated either with the bacterial surface or secreted into the mucosal biofilm matrix, enabling the mucosal layer to bind viruses and impede their access to underlying epithelial cells. Importantly, this may lead to prolonged exposure of viruses to inactivating substances naturally secreted by lactobacilli, such as lactic acid and hydrogen peroxide, thereby significantly reducing the numbers of infectious viral particles.
The expression of heterologous proteins has been achieved in Gram-positive bacteria, including lactobacilli, lactococci, and streptococci (8–11). Although genetic manipulation of a human vaginal isolate Lactobacillus fermentum was reported (12), none has been applied to expression of heterologous mammalian proteins in human vaginal isolates of lactobacilli. In this report, we describe the engineering of a natural colonizing vaginal isolate of L. jensenii to express a secreted form of the prototypical HIV-binding protein, CD4. We demonstrate that 2D CD4 produced by these bacteria exhibit full biological activity in vitro, defined by the ability to bind gp120 and to inhibit HIV-1 viral entry. This work provides an important first step toward the development of engineered lactobacilli, applied onto the vaginal mucosa, to block the sexual transmission of HIV in women.
Materials and Methods
Bacterial Strains and Culture. Naturally occurring human vaginal isolates of L. crispatus, L. gasseri, and L. jensenii, including Xna strain used in this study, were obtained from vaginal swabs of healthy volunteers and routinely cultivated at 37°C (5% CO2/95% air) in either de Man, Rogosa, and Sharpe (MRS) broth or Rogosa SL broth (Difco). For protein expression analysis, Medium 199 (Invitrogen) was also used. Plasmids were introduced by electroporation into Escherichia coli DH12S (Invitrogen). For plasmid maintenance, the transformed E. coli DH12S were grown in LB broth (Difco) at 37°C, supplemented with erythromycin (300 μg/ml). Plasmids were transformed by electroporation into L. jensenii essentially as described for L. gasseri (13). The transformed L. jensenii bacteria were routinely propagated in liquid media containing 20 μg/ml erythromycin.
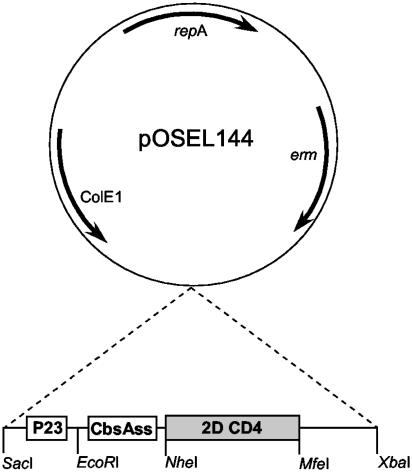
Construction of Expression Vector. Shuttle vector pOSEL144 was created by first subcloning the E. coli origin of replication from pBluescript into pVE5528 (14), and then removing the full-length M6 coding region (PstI). This plasmid was digested with SmaI, partially digested with NdeI, filled-in by DNA polymerase I (Klenow fragment) and self-ligated. The resulting plasmid, pOSEL144 (Fig. 1), contains a lactobacillus-compatible origin of replication (repA) and an erythromycin resistance gene (erm) from pLEM7 (14), and has been used for the expression of heterologous proteins in a variety of lactobacillus species.
Fig. 1.
Schematic of the pOSEL144 expression vector. The repA and erm genes are derived from a naturally occurring L. reuteri plasmid pLEM7 (14) and the ColE1 replicon is from pBluescript. An expression cassette comprising the P23 promoter from Lactococcus lactis (15), the CbsAss from L. crispatus, and the 2D CD4 coding region was inserted into SacI and XbaI sites of pOSEL144. The resulting plasmid was designated as pOSEL651.
To clone HIV-binding ligands for secretion in L. jensenii strain Xna, an expression cassette was constructed by PCR amplification and subcloned into the SacI and XbaI sites of pOSEL144 (Fig. 1). The cassette contains four components, including lactobacillus-compatible promoter elements, signal sequence for secretion, HIV-binding ligands, and cell wall anchoring domain. Unique restriction sites, including SacI, EcoRI, NheI, MfeI, and XbaI were placed between each component from 5′ to 3′ ends, respectively. Amplification of each component by PCR was performed by using Pfu DNA polymerase. The P23 promoter from Lactococcus lactis (15) was created by amplification with 5′-GTGGAGCTCCCCGAAAAGCCCTGACAACCC-3′ and 5′-GGAAACACGCTAGCACTAACTTCATT-3′ primers. To direct secretion of the 2D-CD4 protein, two primers, 5′-GCGAATTCAAGGAGGAAAAGACCACAT-3′ and 5′-CCAGCTAGCTGAAACAGTAGAAACGGC-3′, were designed to amplify the S-layer gene (cbsA) sequence of L. crispatus from the putative ribosome binding site to the signal peptidase cleavage site, with EcoRI and NheI sites added to the 5′ and 3′ ends, respectively (restriction sites are underlined). The amplified S-layer signal nucleotide sequence (CbsAss) corresponding to MKKNLRIVSAAAAALLAVAPVAA was then digested and used for cloning in our expression cassette.
Synthesis and Secretion of 2D CD4 in L. jensenii. The first two extracellular domains (K1–S183) of human CD4 (2D CD4) that is capable of high-affinity binding to gp120 as four-domain sCD4 (16) was recoded by assembly PCR (17) to conform more closely to the optimal lactobacillus codon usage. The 2D CD4 was amplified by 5′-GCGGCTAGCAAGAAAGTTGTTTTAGGTAAA-3′ and 5′-GCACAATTGTGATGCCTTTTGAAAAGCTAA-3′ using the recoded CD4 sequence. All of the resulting PCR products were digested by respective restriction enzymes, and fragments of DNA were ligated with EcoRI/XbaI-digested pOSEL144. In addition, a TAA stop codon was inserted into the N-terminal region of LPXTG anchoring motif to ensure protein secretion. The resulting plasmid was designated as pOSEL651 (Fig. 1). DNA sequencing was conducted for sequence verification before transformation into L. jensenii.
Partial Purification of Lactobacillus-Derived 2D CD4. Culture supernatants in strain Xna-651 or Xna-144 were treated with protease inhibitor mixture (Roche Applied Science), dialyzed against 50 mM sodium phosphate, pH 6.8, at 4°C in a 10-kDa cutoff dialysis membrane, and then passed over a column of Q Sepharose Fast Flow resin (Amersham Biosciences). The 2D CD4 proteins in the flow through were then bound to SP Sepharose Fast Flow (Amersham Biosciences) and eluted in the buffer containing 0.8 M NaCl. Similarly, the mock samples from the control strain Xna-144 as a result of nonspecific binding to ion exchange matrixes were also prepared. The amount of eluted 2D CD4 was determined by Western blot and activity analyzed by CD4 ELISA and gp120 binding using soluble CD4 (AIDS Research and Reference Reagent Program, National Institutes of Health) as a standard.
Western Analysis of 2D CD4 Expression in L. jensenii. Strain Xna-651 or Xna-144 was grown at 37°C and 5% CO2/95% air in Rogosa SL broth buffered with 100 mM Hepes (pH 7.4). Proteins in cell-free conditioned media were precipitated with trichloracetic acid at a final concentration of 20%. The resulting protein precipitates were washed with ethanol, air dried, and heat denatured in SDS/PAGE loading buffer (50 mM Tris·HCl, pH 6.8/10 mM DTT/0.4% SDS/6% sucrose/0.01% bromophenol blue). Afterward, soluble proteins, along with E. coli-derived and refolded 2D CD4 as a standard, were resolved by SDS/PAGE using a 4–12% NuPAGE system (Invitrogen), and then electroblotted on to poly(vinylidene fluoride) membranes. The blot was probed with the polyclonal rabbit anti-CD4 antibody T4-4 (AIDS Research and Reference Reagent Program, National Institutes of Health). The antigen-antibody reaction was then visualized by using alkaline phosphatase-conjugated anti-rabbit IgG and chromogenic detection reagents (Promega).
Expression of 2D CD4 in E. coli and Refolding. PCR amplified 2D CD4 was cloned into NdeI and BamHI sites of pET-11a (Novagen) and subsequently transformed into BL21DE3 (Novagen). The transformed bacterial cells were cultured at 37°C in the presence of 100 μg/ml ampicillin to OD600 = 0.4. After 2-hr induction with 1 mM isopropyl-β-d-galactopyranoside, bacterial cells were harvested, lysed by sonication in the presence of protease inhibitor mixture (Roche Applied Science), and subfractionated by centrifugation. The 2D CD4 containing inclusion bodies were homogenized and washed in 2 mM EDTA, 0.5 M NaCl, 1% (vol/vol) Triton X-100, and 50 mM sodium acetate, pH 6.0, before being solubilized in 8 M guandine hydrochloride, 10 mM EDTA, 10 mM DTT, and 100 mM Tris, pH 7.0, at room temperature. The resulting soluble proteins were added quickly to a ice-cold oxidation and refolding solution containing 50 mM Tris, 1 M urea, 0.25 mM EDTA, 10 mM l-cysteine, and 1 mM cystamine, pH 8.0. The refolding reaction was allowed to proceed for 10 h at 4°C with slow stirring of the solution. Afterward, the refolded 2D CD4 proteins were dialyzed against PBS in 10-kDa cutoff membranes (Spectrum Laboratories). The biological activity of 2D CD4 was quantified by ELISA and gp120 binding using soluble CD4 (AIDS Research and Reference Reagent Program, National Institutes of Health) as a standard.
CD4 ELISA and HIV-1 gp120 Binding Assay. The concentration of correctly folded 2D CD4 proteins was determined by CD4 capture ELISA. Correctly folded 2D CD4 protein secreted into the culture supernatants was captured on microtiter plates by monoclonal antibody Leu3A or Sim.4 at 10 μg/ml (18). Bound 2D CD4 molecules were probed with polyclonal rabbit antibodies, T4-4, then detected by alkaline phosphatase-conjugated anti-rabbit IgG and AMPAK ELISA Amplication system (Dako).
The ability of this 2D CD4 preparation to interact with gp120 was analyzed by gp120 binding assay (19) with modifications. Briefly, 96-well microtiter plates coated with sheep antibody D7324 against HIV-1 gp120 (Aalto Bio, Dublin) at 5 μg/ml were blocked with Tris-buffered saline containing 2% nonfat milk, washed and then incubated with 10 ng per well recombinant gp120 (rgp120, Protein Sciences, Meriden, CT). Samples containing CD4 molecules were then captured by rgp120, and bound CD4 molecules were detected by polyclonal antibodies, T4-4, as described above.
HIV-1 Infection Assay. The ability of 2D CD4 proteins expressed by Xna-651 to inhibit HIV-1 entry was determined by using a single-cycle infection assay (20, 21). To produce HIV-1HxB2 strain Env-pseudotyped, luciferase-expressing reporter viruses, HEK293T cells were cotransfected with a plasmid encoding an envelope-deficient HIV-1 NL4–3 virus, the pNL-Luc plasmid carrying the luciferase reporter gene, and a pSV plasmid expressing viral envelope glycoproteins derived from HIV-1HxB2 strain. Similarly, HIV-1JR-FL Env-pseudotyped, luciferaseexpressing reporter viruses were produced. Subsequently, the virus p24 antigen concentration in virus-containing supernatant was determined by p24 antigen ELISA (Coulter). One day before infection, HeLa-CD4-CXCR4 cells were seeded at 5 × 104 cells per well in a 48-well tissue culture plate. Envpseudotyped reporter HIV-1HxB2 viruses tittered according to Connor et al. (21) were first incubated with different concentrations of 2D CD4 protein (37°C, 30 min), either directly from culture supernatants or prepared after partial purification. Subsequently, the virus in the reaction mixtures was used to infect cells (37°C, 1 h). Unbound virus was removed by washing with PBS.
Alternatively, Xna-651 was resuspended at 2.5 × 108 cells per ml and cultured in Medium 199 (37°C, 30 min). Then, the bacteria, along with input Env-pseudotyped reporter HIV-1HxB2 or HIV-1JR-FL virus tittered according to Connor et al. (21), were added to HeLa cells expressing CD4–CXCR4–CCR5 (22), respectively (37°C, 1 h). Subsequently, cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS (37°C, 48 h, 5% CO2/95% air). The cells were washed once with PBS and lysed with reporter lysis buffer (Promega). Luciferase activity in a mixture of luciferase substrate and cell lysate was measured in relative light units with a Chiron microplate luminometer.
Results
Expression of 2D CD4 in Human Vaginal Isolates of L. jensenii. Multiple human vaginal isolates of L. jensenii, L. crispatus, and L. gasseri (isolated from healthy women of child-bearing age) were evaluated for efficient growth, transformability with foreign DNA, level of functional protein expression, and in vitro cytotoxicity to HeLa cells. Strain Xna of L. jensenii was among a few strains selected for the expression of recombinant CD4 molecules based on these characteristics. To express heterologous proteins in this strain, an expression vector derived from a natural L. reuteri plasmid pVE5528 (14) was constructed by using a modular approach (Fig. 1). Because promoter and N-terminal secretion signal sequences are critical factors affecting the efficiency of transcription and protein expression (23, 24), a series of promoters [P23 or P59 of Lactococcus lactis (15, 25) and a proprietary S-layer protein (cbsA) promoter element], along with secretion signals for M6 protein of Streptococcus pyogenes (25), α-Amylase of L. amylovorus (26), and S-layer protein (CbsA) of L. crispatus were analyzed. A combination of the P23 promoter from Lactococcus lactis and the signal sequence from the CbsA of L. crispatus, as illustrated in Fig. 1, was found to drive the highest levels of 2D CD4 expression.
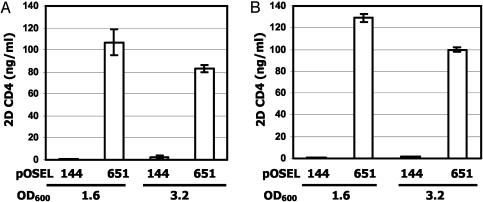
On establishment of electroporation conditions for transforming L. jensenii, the 2D CD4 expression plasmid, pOSEL651, and the control plasmid, pOSEL144 (Fig. 1), were introduced into L. jensenii Xna, yielding strains Xna-651 and Xna-144, respectively. To determine whether Xna-651 expressed 2D CD4 protein, culture supernatants were harvested at late-exponential phase (OD600 = 0.8) and stationary phase (OD600 = 3.2) for analyses of soluble proteins. Western blot analysis using polyclonal anti-CD4 antibodies (T4-4) detected 2D CD4 at the predicted size secreted into the supernatant of Xna-651 at late log phase (Fig. 2). This 2D CD4 protein expressed in modified L. jensenii Xna or 1153 strain was released at concentrations ranging from 100 ng/ml to 1 μg/ml depending on the experimental conditions, when quantified in Western blotting in reference to known concentrations of E. coli-derived 2D CD4 (data not shown). The 2D CD4 was also detected well after active cell division had ceased. This protein was not detected in supernatants of the Xna-144 strain.
Fig. 2.
Time-dependent release of 2D CD4 by L. jensenii Xna harboring the 2D CD4 expression plasmid (Xna-651) relative to the control (Xna-144). Samples were collected at OD600 0.8 or 3.2. Proteins were collected by precipitation with TCA from culture supernatants from corresponding bacterial cells and subjected to electrophoretic separation under reducing SDS/PAGE for Western blot analysis with polyclonal anti-CD4 antibodies, T4-4.
Secretion of Correctly Refolded 2D CD4. To characterize the biological activity of the secreted 2D CD4, ELISA assays were performed (Fig. 3). Polyclonal antibodies against CD4 (T4-4) were used to detect bound 2D CD4 molecules, after their capture onto a solid support by conformation-dependent Leu3A monoclonal antibody or HIV-1 gp120. Functional 2D CD4 (≈100 ng/ml) was detected in the culture supernatant by these assays (Fig. 3). The concentration of functional 2D CD4 remained elevated during extended incubation overnight. Similar quantitative results were obtained with either Leu3A or HIV-1 gp120 to capture 2D CD4. 2D CD4 was not detected in the absence of Leu3A or HIV gp120 capture protein (data not shown).
Fig. 3.
Secretion of biologically active 2D CD4 by engineered L. jensenii Xna. L. jensenii Xna-144 or Xna-651 bacteria were grown in MRS broth (37°C and 5% CO2). Culture supernatants were used for CD4 ELISA (A) and gp120 binding assay (B). 2D CD4 concentrations were determined from standard curves generated with refolded 2D CD4 expressed in E. coli.
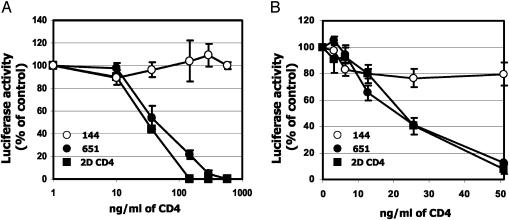
L. jensenii-Derived 2D CD4 Inhibits HIV-1 Infectivity in Vitro. The first two steps in the HIV-1 infectious process are attachment of virus to CD4 on the target cell and the fusion of attached virus with the target cell plasma membrane. To determine whether 2D CD4 secreted from strain Xna-651 blocks infection, we used HIV-1HxB2, carrying a luciferase reporter to infect HeLa cells expressing CD4 and CXCR4 in a single cycle viral infection (20, 21). In initial experiments, soluble 2D CD4, secreted into the culture supernatant by Xna-651, was partially purified by using ion-exchange chromatography. As a positive control, a partially purified and refolded recombinant 2D CD4 preparation, produced in E. coli, was added to samples from the control strain, Xna-144. Protein released from Xna-651 inhibited HIV-1 infectivity in a dose-dependent manner, similar to the positive control, whereas an equal volume of mock samples prepared as described in Materials and Methods from the control strain (Xna-144) had no effect (Fig. 4A).
Fig. 4.
L. jensenii-derived 2D CD4 inhibits infectivity of Env-pseudotyped HIV-1HxB2. The inhibitory effect of the 2D CD4 protein produced by Xna-651 after partial purification (A) and conditioned medium 199 containing 2D CD4 (B) on Env-pseudotyped HIV-1HxB2 infectivity of HeLa cells expressing CD4-CXCR4 was determined by viral entry assay (21). The results (mean ± SD) from triplicate determinations in a single experiment were presented and confirmed in three independent experiments. The luciferase activity from virus-infected cells without addition of 2D CD4 samples was defined as 100%. This corresponded to relative light unit values of 276 ± 21 in A and 350 ± 19 in B.
As a complementary approach, supernatants from Xna-651 or Xna-144 were used directly in neutralization experiments. Conditioned media was prepared by incubating L. jensenii at log phase in a HeLa cell compatible medium, Medium 199. Based on both CD4 ELISA and HIV-1 gp120 binding analysis, biologically active 2D CD4 at 50 ng per 5 × 108 colony-forming unit/ml was detected in the supernatant of Xna-651, but not in supernatants derived from Xna-144. Addition of this Xna-651-conditioned medium to HeLa–CD4 cells inhibited HIV-1 infectivity in a dose-dependent manner, whereas Xna-144-conditioned media was inactive (Fig. 4B). Inhibitory concentrations of Xna-651-derived 2D CD4 were similar to those obtained when E. coli-derived 2D CD4 standard was added to conditioned media of Xna-144.
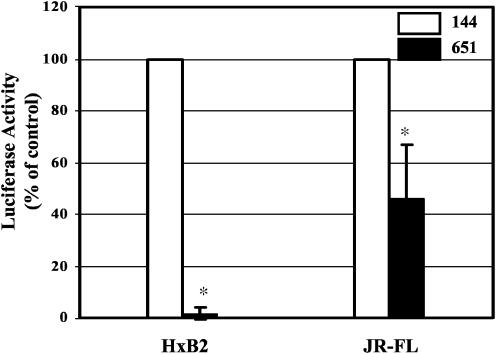
Coincubation of Strain Xna-651 Bacteria with Recombinant HIV-1 Inhibits Viral Entry in Vitro. Primary strains of HIV are known to be less sensitive to inhibition by CD4 than laboratory-adapted strains (27). The primary strain JR-FL has been shown to be at least 50-fold less sensitive to CD4 inhibition as compared with the laboratory-adapted strain HxB2 (27, 28). Single-cycle infection assays using Env-pseudotyped HIV-1HxB2 reporter virus demonstrated that coincubation of L. jensenii Xna-651 with HeLa cells expressing CD4–CXCR4 decreased HIV-1 infectivity in a dose-dependent manner (data not shown). Accordingly, experiments were performed to further evaluate whether coincubation of L. jensenii Xna-651 with pseudotyped HIV-1, containing the envelope from either the laboratory strain HxB2 or the primary strain JR-FL, could decrease virus entry into susceptible cell lines (Fig. 5). L. jensenii Xna-651 and Xna-144 were added along with Env-pseudotyped HIV-1HxB2 virus or Env-pseudotyped HIV-1JR-FL virus to HeLa cells expressing CD4–CXCR4–CCR5 as described in Materials and Methods. Both L. jensenii strains are cytotoxic to HeLa cells in cell cultures at numbers >2.5 × 108 bacteria per ml, which limited the number of bacteria used in these experiments. Despite this limitation, the Xna-651 strain exhibited modest but statistically significant inhibitory effects on HIV-1 entry against both pseudotypes via virus-associated luciferase activity at 48 h after infection (Fig. 5). Under these conditions, Xna-651 inhibited HIV-1HxB2 entry by 95% (P < 0.01) and HIV-1JR-FL entry by 55% (P < 0.01), consistent with the differential sensitivity to CD4 of the two HIV strains, relative to the control lactobacillus Xna-144 at the same cell density of 2.5 × 108 bacteria per ml. These results demonstrate clearly that a natural human isolate of L. jensenii, engineered to express 2D CD4, inhibits the infection of cultured cells by a recombinant laboratory-adapted, and moderately inhibits a primary, strain of HIV-1, thereby providing an in vitro demonstration of this anti-infective strategy.
Fig. 5.
Coincubation of L. jensenii Xna-651 bacteria with HeLa-CD4 cells and recombinant viruses inhibits HIV-1 infectivity. L. jensenii Xna-144 or Xna-651 bacteria in medium 199 were coincubated with Env-pseudotyped HIV-1HxB2 or HIV-1JR-FL viruses and HeLa-CD4-CCR5 for 1 h at 37°C in the viral entry assay (21). The results (mean ± SD) from four experiments are presented. The luciferase activity from virus-infected cells in the presence of Xna-144 was defined as 100%. The statistical significance of the data (*, P < 0.01) was determined by Student's t test.
Discussion
Numerous avenues are currently being investigated to curtail the HIV epidemic (29–32). Although an effective HIV vaccine remains the most important goal, the ability of HIV to mutate rapidly and evade the immune response has made the development of a vaccine problematic, necessitating the pursuit of additional innovative approaches. Topical microbicides represent a possible approach to inhibit the transmission of sexually transmitted disease pathogens, including HIV, in women. However, most microbicides currently under development act via nonspecific mechanisms, with a risk of irritating underlying vaginal tissue and thereby paradoxically increasing the risk of infection, as was shown with nonoxynol-9 (33). We propose a unique mode of impeding the transmission of HIV through a receptor-ligand specific mechanism. This approach involves the administration of naturally colonizing commensal lactobacilli engineered to express proteins for the purpose of neutralizing pathogenic viral particles within the mucosal microflora. The application of this technology could impede the transmigration of viral particles within the mucosal microbial biofilm of the vagina and diminish their access to receptor sites on host cells and tissues.
We have obtained natural human isolates of L. jensenii and adapted methods for the genetic manipulation of these commensal bacteria. In this paper, we demonstrate the expression of a prototypical HIV-binding protein, 2D CD4 (34), by a carefully selected strain (L. jensenii Xna) exhibiting favorable growth and colonization properties. Our studies demonstrate that 2D CD4 is expressed by L. jensenii strain Xna-651 in a native conformation based on immunoreactivity with the conformational dependent antibodies Leu3A or Sim.4, binding to HIV-1 gp120, and inhibition of HIV-1 infectivity in vitro. These findings demonstrate that a human vaginal isolate of L. jensenii is capable of expressing a complex, disulfide-bonded protein in a biologically active form.
Our data demonstrate that 2D CD4 proteins secreted by L. jensenii are capable of inhibiting isolates of HIV-1 as judged by several assay formats. The observation that the inhibitory concentrations of bacterially expressed 2D CD4 in spent culture media are similar to those of a partially purified refolded 2D CD4 standard produced in E. coli provides compelling evidence that the 2D CD4 molecule itself mediates the inhibition of HIV-1 infectivity. The significance of this result is further enhanced by the use of conditions that mimic the natural situation through the use of an assay demonstrating that engineered lactobacilli exhibit the inhibitory activity when coincubated directly with recombinant HIV-1 viruses pseudotyped with envelopes from laboratory-adapted X4 or primary R5 strains. Entry of Envpseudotyped HIV-1HxB2 virus (a laboratory strain) and Env-pseudotyped HIV-1JR-FL (a primary clinical strain) were both inhibited (P < 0.01) after incubation with modified lactobacilli. The greater inhibition of the pseudotyped laboratory strain of HIV-1 (HIV-1HxB2) compared with the pseudotyped primary strain (HIV-1JR-FL) is consistent with the reduced sensitivity of primary strains to CD4 as compared with laboratory strains, and is further confirmation of the preserved biological activity of CD4 secreted by engineered lactobacilli. Our current proof-of-concept experiments involve the use of modified L. jensenii harboring plasmids that contain antibiotic resistance markers. Because these plasmids might be transferred to opportunistic human pathogens such as Staphylococci and Enterococci (35), these modified bacteria are not intended for clinical testing. In an important step toward clinical development of engineered L. jensenii, we recently identified sequences for site-specific chromosome integration by homologous recombination and succeeded at stably integrating the gene encoding 2D CD4 into L. jensenii chromosome while the erythromycin resistance gene was excised (data not shown).
Our strategy envisions the intravaginal administration of engineered L. jensenii to women as a means of blocking HIV transmission. Although initial proof-of-principle was demonstrated by using the prototypical HIV ligand CD4 in this report, other proteins exist that could more efficiently bind and neutralize HIV within the mucosa, such as broadly neutralizing antibodies, CD4-antibody fusion proteins, or cyanovirin-N (36–38). Other promising candidates include variants of CD4 capable of forming oligomers that exhibit an enhanced avidity for binding primary isolates of HIV-1 (39, 40). Our technology may be used to engineered lactobacilli to secrete such proteins. Alternatively, CD4 or other HIV-binding proteins may be covalently anchored to the cell wall of L. jensenii as an approach to trapping and immobilizing virions on the bacterial surface. Once captured in this manner, the viral particles are likely to be rendered noninfectious by exposure to locally produced virocidal substances such as lactic acid and hydrogen peroxide. As a result, the number of infectious viral particles is likely to be reduced. Because the cervico-vaginal transmission of HIV is already an inefficient process, a reduction of the infectious viral load should further reduce transmission frequencies (41).
An important element in this concept is the persistence and colonization of the engineered lactobacilli on the vaginal mucosa. Although this remains to be tested in vivo, expression of 2D CD4 did not appear to affect expression of cell surface-associated proteins in modified bacteria, when probed with membrane-impermeable sulfo-N-hydroxysuccinimide-biotin according to Sabarth et al. (42), nor the adherence of modified bacteria to protein matrices (data not shown). Importantly, a growing body of evidences that modified Gram-positive bacteria or exogenously applied lactobacilli can colonize and persist in relevant animal models or women already exists. For instance, recombinant strains of Streptococcus gordonii or Lactobacillus zeae secreting or surface displaying single-chain antibody stably colonize in rat vaginal or oral mucosa (9, 10). Furthermore, Hillier et al. demonstrated that human isolates of L. crispatus, inoculated intravaginally in women with bacterial vaginosis, persist for periods of weeks to months (S. Hillier, personal communication). This and the above-mentioned observations support the notion that carefully selected, human-derived lactobacilli genetically modified to express CD4, or related HIV-binding proteins, may persist on the vaginal mucosa for a reasonable period, perhaps days to weeks. If this proves to be true, then such bacteria could be administrated intermittently (possibly once or twice weekly) to provide prolonged, ongoing protection against HIV infection. This important feature further differentiates our approach from other sexually transmitted disease preventatives, such as condoms and chemical microbicides, which require application immediately preceding each potential exposure. The findings reported in this paper represent an important step toward the development of a much-needed discrete, female-controlled intervention against HIV transmission.
Acknowledgments
We thank Drs. Shawn Kuhmann and James Binley for technical advice, and Drs. Edward Berger and Mario Roederer for critical reading of this manuscript. This work was funded by National Institutes of Health Small Business Innovation Research Phase I Grant AI-46203-01 and by the Global Microbicide Project (GMP-01-09) of the Contraceptive Research and Development Program, Eastern Virginia Medical School (Norfolk).
Abbreviations: HIV-1, HIV type 1; 2D CD4, two-domain human CD4; CbsA, S-layer protein of Lactobacillus crispatus.
References
- 1.Royce, R. A., Sena, A., Cates, W., Jr., & Cohen, M. S. (1997) N. Engl. J. Med. 336, 1072–1078. [DOI] [PubMed] [Google Scholar]
- 2.al-Nozha, M., Ramia, S., al-Frayh, A. & Arif, M. (1990) J. Acquired Immune Defic. Syndr. 3, 193–194. [PubMed] [Google Scholar]
- 3.Redondo-Lopez, V., Cook, R. L. & Sobel, J. D. (1990) Rev. Infect. Dis. 12, 856–872. [DOI] [PubMed] [Google Scholar]
- 4.Taha, T. E., Hoover, D. R., Dallabetta, G. A., Kumwenda, N. I., Mtimavalye, L. A., Yang, L. P., Liomba, G. N., Broadhead, R. L., Chiphangwi, J. D. & Miotti, P. G. (1998) AIDS 12, 1699–1706. [DOI] [PubMed] [Google Scholar]
- 5.Antonio, M. A., Hawes, S. E. & Hillier, S. L. (1999) J. Infect. Dis. 180, 1950–1956. [DOI] [PubMed] [Google Scholar]
- 6.Pavlova, S. I., Kilic, A. O., Kilic, S. S., So, J.-S., Nader-Macias, M. E., Simoes, J. A. & Tao, L. (2002) J. Appl. Microbiol. 92, 451–459. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., Stewart, P. S. & Greenberg, E. P. (1999) Science 284, 1318–1322. [DOI] [PubMed] [Google Scholar]
- 8.Steidler, L., Hans, W., Schotte, L., Neirynck, S., Obermeier, F., Falk, W., Fiers, W. & Remaut, E. (2000) Science 289, 1352–1355. [DOI] [PubMed] [Google Scholar]
- 9.Beninati, C., Oggioni, M. R., Boccanera, M., Spinosa, M. R., Maggi, T., Conti, S., Magliani, W., De Bernardis, F., Teti, G., Cassone, A., et al. (2000) Nat. Biotechnol. 18, 1060–1064. [DOI] [PubMed] [Google Scholar]
- 10.Kruger, C., Hu, Y., Pan, Q., Marcotte, H., Hultberg, A., Delwar, D., Van Dalen, P. J., Pouwels, P. H., Leer, R. J., Kelly, C. G., et al. (2002) Nat. Biotechnol. 20, 702–706. [DOI] [PubMed] [Google Scholar]
- 11.Giomarelli, B., Provvedi, R., Meacci, F., Maggi, T., Medaglini, D., Pozzi, G., Mori, T., McMahon, J. B., Gardella, R. & Boyd, M. R. (2002) AIDS 16, 1351–1356. [DOI] [PubMed] [Google Scholar]
- 12.Pavlova, S. I., Kilic, A. O., Topisirovic, L., Miladinov, N., Hatzos, C. & Tao, L. (2002) Plasmid 47, 182–192. [DOI] [PubMed] [Google Scholar]
- 13.Luchansky, J. B., Tennant, M. C. &Klaenhammer, T. R. (1991) J. Dairy Sci. 74, 3293–3302. [DOI] [PubMed] [Google Scholar]
- 14.Fons, M., Hege, T., Ladire, M., Raibaud, P., Ducluzeau, R. & Maguin, E. (1997) Plasmid 37, 199–203. [DOI] [PubMed] [Google Scholar]
- 15.van der Vossen, J. M., van der Lelie, D. & Venema, G. (1987) Appl. Environ. Microbiol. 53, 2452–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salzwedel, K., Smith, E. D., Dey, B. & Berger, E. A. (2000) J. Virol. 74, 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stemmer, W. P., Crameri, A., Ha, K. D., Brennan, T. M. & Heyneker, H. L. (1995) Gene 164, 49–53. [DOI] [PubMed] [Google Scholar]
- 18.McCallus, D. E., Ugen, K. E., Sato, A. I., Williams, W. V. & Weiner, D. B. (1992) Viral. Immunol. 5, 163–172. [DOI] [PubMed] [Google Scholar]
- 19.Moore, J. P. (1993) AIDS Res. Hum. Retroviruses 9, 209–219. [DOI] [PubMed] [Google Scholar]
- 20.Chen, B. K., Saksela, K., Andino, R. & Baltimore, D. (1994) J. Virol. 68, 654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connor, R. I., Sheridan, K. E., Ceradini, D., Choe, S. & Landau, N. R. (1997) J. Exp. Med. 185, 621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Platt, E. J., Wehrly, K., Kuhmann, S. E., Chesebro, B. & Kabat, D. (1998) J. Virol. 72, 2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCracken, A. & Timms, P. (1999) J. Bacteriol. 181, 6569–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dieye, Y., Usai, S., Clier, F., Gruss, A. & Piard, J. C. (2001) J. Bacteriol. 183, 4157–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piard, J. C., Hautefort, I., Fischetti, V. A., Ehrlich, S. D., Fons, M. & Gruss, A. (1997) J. Bacteriol. 179, 3068–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitzsimons, A., Hols, P., Jore, J., Leer, R. J., O'Connell, M. & Delcour, J. (1994) Appl. Environ. Microbiol. 60, 3529–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daar, E., Li, X., Moudgil, T. & Ho, D. (1990) Proc. Natl. Acad. Sci. USA 87, 6574–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKeating, J. A., Bennett, J., Zolla-Pazner, S., Schutten, M., Ashelford, S., Brown, A. L. & Balfe, P. (1993) J. Virol. 67, 5216–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Root, M. J., Kay, M. S. & Kim, P. S. (2001) Science 291, 884–888. [DOI] [PubMed] [Google Scholar]
- 30.Jones, D. L., Weiss, S. M., Malow, R., Ishii, M., Devieux, J., Stanley, H., Cassells, A., Tobin, J. N., Brondolo, E., LaPerriere, A., et al. (2001) J. Urban Health 78, 593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letvin, N. L. (2002) J. Clin. Invest. 110, 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veazey, R. S., Shattock, R. J., Pope, M., Kirijan, J. C., Jones, J., Hu, Q., Ketas, T., Marx, P. A., Klasse, P. J., Burton, D. R., et al. (2003) Nat. Med. 9, 343–346. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson, D., Tholandi, M., Ramjee, G. & Rutherford, G. W. (2002) Lancet Infect. Dis. 2, 613–617. [DOI] [PubMed] [Google Scholar]
- 34.Ryu, S. E., Kwong, P. D., Truneh, A., Porter, T. G., Arthos, J., Rosenberg, M., Dai, X. P., Xuong, N. H., Axel, R., Sweet, R. W., et al. (1990) Nature 348, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teuber, M., Meile, L. & Schwarz, F. (1999) Antonie Van Leeuwenhoek 76, 115–137. [PubMed] [Google Scholar]
- 36.Pantophlet, R., Ollmann Saphire, E., Poignard, P., Parren, P. W., Wilson, I. A. & Burton, D. R. (2003) J. Virol. 77, 642–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dey, B., Del Castillo, C. S. & Berger, E. A. (2003) J. Virol. 77, 2859–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyd, M., Gustafson, K., McMahon, J., Shoemaker, R., O'Keefe, B., Mori, T., Gulakowski, R., Wu, L., Rivera, M., Laurencot, C., et al. (1997) Antimicrob. Agents Chemother. 41, 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arthos, J., Cicala, C., Steenbeke, T. D., Chun, T.-W., Cruz, C. D., Hanback, D. B., Khazanie, P., Nam, D., Schuck, P., Selig, S. M., et al. (2002) J. Biol. Chem. 277, 11456–11464. [DOI] [PubMed] [Google Scholar]
- 40.Kwong, P. D., Doyle, M. L., Casper, D. J., Cicala, C., Leavitt, S. A., Majeed, S., Steenbeke, T. D., Venturi, M., Chaiken, I., Fung, M., et al. (2002) Nature 420, 678–682. [DOI] [PubMed] [Google Scholar]
- 41.Chakraborty, H., Sen, P. K., Helms, R. W., Vernazza, P. L., Fiscus, S. A., Eron, J. J., Patterson, B. K., Coombs, R. W., Krieger, J. N. & Cohen, M. S. (2001) AIDS 15, 621–627. [DOI] [PubMed] [Google Scholar]
- 42.Sabarth, N., Lamer, S., Zimny-Arndt, U., Jungblut, P. R., Meyer, T. F. & Bumann, D. (2002) J. Biol. Chem. 277, 27896–27902. [DOI] [PubMed] [Google Scholar]